You are viewing the Initial Version, the most recent version of this dataset.
1 version(s) available
Date of publication: October 23, 2024
Version 1
Date of publication: October 23, 2024
Type of change:
Description:
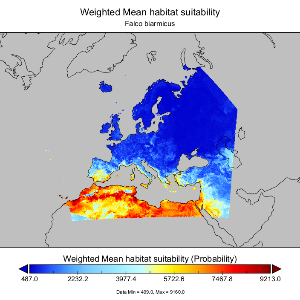
Species habitat suitability of European terrestrial vertebrates for contemporary climate and land use
by Sara Si-MoussiPredicted habitat suitability of most terrestrial vertebrate species (1,207 species) in Europe at 1km resolution under current conditions (1990-2020). Predictions for each species were obtained from an ensemble of species distribution models (ensemble SDMs). The provided data are the raw weighted mean of habitat suitability through the ensemble SDMs, the rescaled weighted mean of habitat suitability through the ensemble SDMs but spatially constra ...(continue reading)
DOI 10.25829/wpfn43Citation
Si-moussi, S., Thuiller, W. (2024). Species habitat suitability of European terrestrial vertebrates for contemporary climate and land use (Version 1) [Dataset]. German Centre for Integrative Biodiversity Research. https://doi.org/10.25829/wpfn43Terrestrial VertebratesEurope UnionSpecies DistributionsPredicted Distributions
| phylum | class | order | family | genus | species | acceptedUsageKey |
|---|---|---|---|---|---|---|
| Chordata | Amphibia | Anura | Alytidae | Alytes | Alytes cisternasii | 2426610 |
| Chordata | Amphibia | Anura | Bufonidae | Bufo | Bufo verrucosissimus | 5217159 |
| Chordata | Amphibia | Anura | Bufonidae | Bufotes | Bufotes balearicus | 10779794 |
| Chordata | Amphibia | Anura | Bufonidae | Bufotes | Bufotes boulengeri | 10400305 |
| Chordata | Amphibia | Caudata | Salamandridae | Triturus | Triturus macedonicus | 2431888 |
| Chordata | Amphibia | Anura | Bufonidae | Bufotes | Bufotes viridis | 9529916 |
| Chordata | Amphibia | Caudata | Salamandridae | Chioglossa | Chioglossa lusitanica | 2431938 |
| Chordata | Amphibia | Anura | Alytidae | Discoglossus | Discoglossus galganoi | 2426616 |
| Chordata | Amphibia | Anura | Alytidae | Discoglossus | Discoglossus jeanneae | 2426620 |
| Chordata | Amphibia | Anura | Alytidae | Discoglossus | Discoglossus montalentii | 2426621 |
| Chordata | Amphibia | Anura | Alytidae | Discoglossus | Discoglossus pictus | 2426617 |
| Chordata | Amphibia | Anura | Alytidae | Discoglossus | Discoglossus sardus | 2426615 |
| Chordata | Amphibia | Caudata | Salamandridae | Calotriton | Calotriton asper | 2431824 |
| Chordata | Amphibia | Caudata | Salamandridae | Euproctus | Euproctus montanus | 2431940 |
| Chordata | Amphibia | Anura | Alytidae | Alytes | Alytes dickhilleni | 2426613 |
| Chordata | Amphibia | Caudata | Salamandridae | Euproctus | Euproctus platycephalus | 2431941 |
| Chordata | Amphibia | Anura | Hylidae | Hyla | Hyla arborea | 2427573 |
| Chordata | Amphibia | Anura | Hylidae | Hyla | Hyla intermedia | 2427598 |
| Chordata | Amphibia | Anura | Hylidae | Hyla | Hyla meridionalis | 2427555 |
| Chordata | Amphibia | Anura | Hylidae | Hyla | Hyla sarda | 2427563 |
| Chordata | Amphibia | Anura | Hylidae | Hyla | Hyla savignyi | 2427599 |
| Chordata | Amphibia | Caudata | Salamandridae | Lyciasalamandra | Lyciasalamandra antalyana | 2431840 |
| Chordata | Amphibia | Caudata | Salamandridae | Lyciasalamandra | Lyciasalamandra atifi | 2431832 |
| Chordata | Amphibia | Caudata | Salamandridae | Lyciasalamandra | Lyciasalamandra billae | 2431830 |
| Chordata | Amphibia | Caudata | Salamandridae | Lyciasalamandra | Lyciasalamandra fazilae | 2431839 |
| Chordata | Amphibia | Anura | Alytidae | Alytes | Alytes muletensis | 2426609 |
| Chordata | Amphibia | Caudata | Salamandridae | Lyciasalamandra | Lyciasalamandra flavimembris | 2431831 |
| Chordata | Amphibia | Caudata | Salamandridae | Lyciasalamandra | Lyciasalamandra helverseni | 2431829 |
| Chordata | Amphibia | Caudata | Salamandridae | Lyciasalamandra | Lyciasalamandra luschani | 2431833 |
| Chordata | Amphibia | Caudata | Salamandridae | Mertensiella | Mertensiella caucasica | 2431943 |
| Chordata | Amphibia | Caudata | Salamandridae | Neurergus | Neurergus crocatus | 2431869 |
| Chordata | Amphibia | Caudata | Salamandridae | Neurergus | Neurergus strauchii | 2431868 |
| Chordata | Amphibia | Anura | Pelobatidae | Pelobates | Pelobates cultripes | 2430565 |
| Chordata | Amphibia | Anura | Pelobatidae | Pelobates | Pelobates fuscus | 2430567 |
| Chordata | Amphibia | Anura | Pelobatidae | Pelobates | Pelobates syriacus | 2430566 |
| Chordata | Amphibia | Anura | Pelodytidae | Pelodytes | Pelodytes caucasicus | 5217374 |
| Chordata | Amphibia | Anura | Alytidae | Alytes | Alytes obstetricans | 2426612 |
| Chordata | Amphibia | Anura | Pelodytidae | Pelodytes | Pelodytes ibericus | 2424292 |
| Chordata | Amphibia | Anura | Pelodytidae | Pelodytes | Pelodytes punctatus | 2424293 |
| Chordata | Amphibia | Caudata | Salamandridae | Pleurodeles | Pleurodeles waltl | 2431875 |
| Chordata | Amphibia | Caudata | Proteidae | Proteus | Proteus anguinus | 2432042 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana arvalis | 2426789 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax bedriagae | 2426640 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax bergeri | 2426665 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax cerigensis | 2426670 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax cretensis | 2426672 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana dalmatina | 2426760 |
| Chordata | Amphibia | Anura | Bombinatoridae | Bombina | Bombina bombina | 2423443 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax epeiroticus | 2426648 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana graeca | 2426795 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana iberica | 2426774 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana italica | 2426815 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax kurtmuelleri | 2426650 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana latastei | 2426788 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax lessonae | 2426654 |
| Chordata | Amphibia | Anura | Bombinatoridae | Bombina | Bombina pachypus | 2423436 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana macrocnemis | 2426798 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax perezi | 2426658 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana pyrenaica | 2426793 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax ridibundus | 2426661 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax shqipericus | 2426638 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana tavasensis | 2426770 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana temporaria | 2426805 |
| Chordata | Amphibia | Caudata | Salamandridae | Salamandra | Salamandra atra | 2431781 |
| Chordata | Amphibia | Caudata | Salamandridae | Salamandra | Salamandra corsica | 2431779 |
| Chordata | Amphibia | Caudata | Salamandridae | Salamandra | Salamandra infraimmaculata | 2431777 |
| Chordata | Amphibia | Anura | Bombinatoridae | Bombina | Bombina variegata | 2423435 |
| Chordata | Amphibia | Caudata | Salamandridae | Salamandra | Salamandra lanzai | 2431780 |
| Chordata | Amphibia | Caudata | Salamandridae | Salamandra | Salamandra salamandra | 2431776 |
| Chordata | Amphibia | Caudata | Hynobiidae | Salamandrella | Salamandrella keyserlingii | 5218334 |
| Chordata | Amphibia | Caudata | Salamandridae | Salamandrina | Salamandrina perspicillata | 2431851 |
| Chordata | Amphibia | Caudata | Salamandridae | Salamandrina | Salamandrina terdigitata | 2431852 |
| Chordata | Amphibia | Caudata | Plethodontidae | Speleomantes | Speleomantes ambrosii | 2431455 |
| Chordata | Amphibia | Caudata | Plethodontidae | Speleomantes | Speleomantes flavus | 2431457 |
| Chordata | Amphibia | Caudata | Plethodontidae | Speleomantes | Speleomantes genei | 2431672 |
| Chordata | Amphibia | Caudata | Plethodontidae | Speleomantes | Speleomantes imperialis | 2431452 |
| Chordata | Amphibia | Caudata | Plethodontidae | Speleomantes | Speleomantes italicus | 2431456 |
| Chordata | Amphibia | Anura | Bufonidae | Bufo | Bufo bufo | 5217160 |
| Chordata | Amphibia | Caudata | Plethodontidae | Speleomantes | Speleomantes sarrabusensis | 2431454 |
| Chordata | Amphibia | Caudata | Plethodontidae | Speleomantes | Speleomantes strinatii | 2431453 |
| Chordata | Amphibia | Caudata | Plethodontidae | Speleomantes | Speleomantes supramontis | 2431451 |
| Chordata | Amphibia | Caudata | Salamandridae | Ichthyosaura | Ichthyosaura alpestris | 2431783 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton boscai | 5218408 |
| Chordata | Amphibia | Caudata | Salamandridae | Triturus | Triturus carnifex | 2431887 |
| Chordata | Amphibia | Caudata | Salamandridae | Triturus | Triturus cristatus | 2431885 |
| Chordata | Amphibia | Caudata | Salamandridae | Triturus | Triturus dobrogicus | 2431891 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton helveticus | 5218410 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton italicus | 5218406 |
| Chordata | Amphibia | Anura | Bufonidae | Epidalea | Epidalea calamita | 2422507 |
| Chordata | Amphibia | Caudata | Salamandridae | Triturus | Triturus karelinii | 2431890 |
| Chordata | Amphibia | Caudata | Salamandridae | Triturus | Triturus marmoratus | 2431886 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton montandoni | 5218409 |
| Chordata | Amphibia | Caudata | Salamandridae | Triturus | Triturus pygmaeus | 2431889 |
| Chordata | Amphibia | Caudata | Salamandridae | Ommatotriton | Ommatotriton vittatus | 2431811 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton vulgaris | 5218405 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax caralitanus | 2426644 |
| Chordata | Amphibia | Caudata | Salamandridae | Ommatotriton | Ommatotriton ophryticus | 2431813 |
| Chordata | Amphibia | Caudata | Salamandridae | Calotriton | Calotriton arnoldi | 2431827 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton graecus | 8808626 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton lantzi | 10526275 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton maltzani | 10716997 |
| Chordata | Amphibia | Caudata | Salamandridae | Lissotriton | Lissotriton schmidtleri | 10701330 |
| Chordata | Amphibia | Caudata | Salamandridae | Triturus | Triturus ivanbureschi | 8532266 |
| Chordata | Amphibia | Anura | Alytidae | Alytes | Alytes almogavarii | 10803854 |
| Chordata | Amphibia | Anura | Pelobatidae | Pelobates | Pelobates balcanicus | 10444550 |
| Chordata | Amphibia | Anura | Pelobatidae | Pelobates | Pelobates vespertinus | 10635361 |
| Chordata | Amphibia | Anura | Pelodytidae | Pelodytes | Pelodytes atlanticus | 9391920 |
| Chordata | Amphibia | Anura | Pelodytidae | Pelodytes | Pelodytes hespericus | 9568260 |
| Chordata | Amphibia | Anura | Bufonidae | Bufo | Bufo spinosus | 9421513 |
| Chordata | Amphibia | Anura | Bufonidae | Bufotes | Bufotes cypriensis | 10677799 |
| Chordata | Amphibia | Anura | Bufonidae | Bufotes | Bufotes sitibundus | 10898216 |
| Chordata | Amphibia | Anura | Hylidae | Hyla | Hyla molleri | 2427580 |
| Chordata | Amphibia | Anura | Hylidae | Hyla | Hyla orientalis | 2427565 |
| Chordata | Amphibia | Anura | Hylidae | Hyla | Hyla perrini | 10884026 |
| Chordata | Amphibia | Anura | Ranidae | Pelophylax | Pelophylax cypriensis | 9993678 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana holtzi | 2426798 |
| Chordata | Amphibia | Anura | Ranidae | Rana | Rana parvipalmata | 11691106 |
| Chordata | Aves | Gaviiformes | Gaviidae | Gavia | Gavia stellata | 2481958 |
| Chordata | Aves | Procellariiformes | Procellariidae | Fulmarus | Fulmarus glacialis | 2481433 |
| Chordata | Aves | Accipitriformes | Accipitridae | Accipiter | Accipiter brevipes | 2480578 |
| Chordata | Aves | Accipitriformes | Accipitridae | Accipiter | Accipiter nisus | 2480637 |
| Chordata | Aves | Accipitriformes | Accipitridae | Accipiter | Accipiter gentilis | 2480589 |
| Chordata | Aves | Accipitriformes | Accipitridae | Buteo | Buteo buteo | 2480537 |
| Chordata | Aves | Accipitriformes | Accipitridae | Buteo | Buteo rufinus | 2480564 |
| Chordata | Aves | Accipitriformes | Accipitridae | Buteo | Buteo lagopus | 2480524 |
| Chordata | Aves | Accipitriformes | Accipitridae | Aquila | Clanga pomarina | 2480505 |
| Chordata | Aves | Accipitriformes | Accipitridae | Aquila | Clanga clanga | 2480509 |
| Chordata | Aves | Accipitriformes | Accipitridae | Aquila | Aquila nipalensis | 2480513 |
| Chordata | Aves | Accipitriformes | Accipitridae | Aquila | Aquila adalberti | 2480510 |
| Chordata | Aves | Accipitriformes | Accipitridae | Aquila | Aquila heliaca | 2480500 |
| Chordata | Aves | Accipitriformes | Accipitridae | Aquila | Aquila chrysaetos | 2480506 |
| Chordata | Aves | Accipitriformes | Accipitridae | Aquila | Aquila fasciata | 5844449 |
| Chordata | Aves | Accipitriformes | Accipitridae | Hieraaetus | Hieraaetus pennatus | 2480685 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco naumanni | 9584698 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco tinnunculus | 9616058 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco vespertinus | 2481062 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco eleonorae | 2481037 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco columbarius | 9813242 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco subbuteo | 2481035 |
| Chordata | Aves | Procellariiformes | Procellariidae | Pterodroma | Pterodroma madeira | 2481463 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco biarmicus | 8014966 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco cherrug | 2481034 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco rusticolus | 8069880 |
| Chordata | Aves | Falconiformes | Falconidae | Falco | Falco peregrinus | 2481047 |
| Chordata | Aves | Galliformes | Phasianidae | Lagopus | Lagopus lagopus | 2473421 |
| Chordata | Aves | Galliformes | Phasianidae | Lagopus | Lagopus muta | 5227679 |
| Chordata | Aves | Galliformes | Phasianidae | Tetrao | Tetrao urogallus | 2473577 |
| Chordata | Aves | Galliformes | Phasianidae | Lyrurus | Tetrao tetrix | 7191070 |
| Chordata | Aves | Galliformes | Phasianidae | Lyrurus | Tetrao mlokosiewiczi | 2473680 |
| Chordata | Aves | Galliformes | Phasianidae | Tetrastes | Tetrastes bonasia | 2473663 |
| Chordata | Aves | Procellariiformes | Procellariidae | Bulweria | Bulweria bulwerii | 2481606 |
| Chordata | Aves | Galliformes | Phasianidae | Tetraogallus | Tetraogallus caucasicus | 2473446 |
| Chordata | Aves | Galliformes | Phasianidae | Tetraogallus | Tetraogallus caspius | 2473442 |
| Chordata | Aves | Galliformes | Phasianidae | Alectoris | Alectoris graeca | 2474055 |
| Chordata | Aves | Galliformes | Phasianidae | Alectoris | Alectoris chukar | 2474029 |
| Chordata | Aves | Galliformes | Phasianidae | Alectoris | Alectoris barbara | 2474045 |
| Chordata | Aves | Galliformes | Phasianidae | Alectoris | Alectoris rufa | 2474051 |
| Chordata | Aves | Galliformes | Phasianidae | Ammoperdix | Ammoperdix griseogularis | 2473732 |
| Chordata | Aves | Galliformes | Phasianidae | Francolinus | Francolinus francolinus | 5227919 |
| Chordata | Aves | Galliformes | Phasianidae | Perdix | Perdix perdix | 2473958 |
| Chordata | Aves | Galliformes | Phasianidae | Coturnix | Coturnix coturnix | 2474156 |
| Chordata | Aves | Procellariiformes | Procellariidae | Calonectris | Calonectris diomedea | 2481521 |
| Chordata | Aves | Galliformes | Phasianidae | Phasianus | Phasianus colchicus | 9752149 |
| Chordata | Aves | Charadriiformes | Turnicidae | Turnix | Turnix sylvaticus | 2475001 |
| Chordata | Aves | Gruiformes | Gruidae | Anthropoides | Anthropoides virgo | 2474936 |
| Chordata | Aves | Gruiformes | Gruidae | Grus | Grus grus | 2474950 |
| Chordata | Aves | Gruiformes | Rallidae | Rallus | Rallus aquaticus | 2474831 |
| Chordata | Aves | Gruiformes | Rallidae | Crex | Crex crex | 4408498 |
| Chordata | Aves | Gruiformes | Rallidae | Porzana | Zapornia parva | 2474627 |
| Chordata | Aves | Gruiformes | Rallidae | Porzana | Zapornia pusilla | 2474628 |
| Chordata | Aves | Gruiformes | Rallidae | Porzana | Porzana porzana | 2474621 |
| Chordata | Aves | Gruiformes | Rallidae | Porphyrio | Porphyrio porphyrio | 2474416 |
| Chordata | Aves | Procellariiformes | Procellariidae | Puffinus | Puffinus puffinus | 5229380 |
| Chordata | Aves | Gruiformes | Rallidae | Gallinula | Gallinula chloropus | 5228199 |
| Chordata | Aves | Gruiformes | Rallidae | Fulica | Fulica cristata | 2474381 |
| Chordata | Aves | Gruiformes | Rallidae | Fulica | Fulica atra | 2474377 |
| Chordata | Aves | Otidiformes | Otididae | Otis | Otis tarda | 2474921 |
| Chordata | Aves | Otidiformes | Otididae | Tetrax | Tetrax tetrax | 5228228 |
| Chordata | Aves | Charadriiformes | Haematopodidae | Haematopus | Haematopus ostralegus | 7788295 |
| Chordata | Aves | Charadriiformes | Recurvirostridae | Himantopus | Himantopus himantopus | 5229126 |
| Chordata | Aves | Charadriiformes | Recurvirostridae | Recurvirostra | Recurvirostra avosetta | 2480259 |
| Chordata | Aves | Charadriiformes | Burhinidae | Burhinus | Burhinus oedicnemus | 2482100 |
| Chordata | Aves | Charadriiformes | Glareolidae | Glareola | Glareola pratincola | 2480757 |
| Chordata | Aves | Procellariiformes | Procellariidae | Puffinus | Puffinus yelkouan | 5229377 |
| Chordata | Aves | Charadriiformes | Glareolidae | Glareola | Glareola nordmanni | 2480758 |
| Chordata | Aves | Charadriiformes | Charadriidae | Vanellus | Vanellus vanellus | 2480242 |
| Chordata | Aves | Charadriiformes | Charadriidae | Vanellus | Vanellus spinosus | 5229142 |
| Chordata | Aves | Charadriiformes | Charadriidae | Vanellus | Vanellus indicus | 5229131 |
| Chordata | Aves | Charadriiformes | Charadriidae | Vanellus | Vanellus leucurus | 5229147 |
| Chordata | Aves | Charadriiformes | Charadriidae | Pluvialis | Pluvialis apricaria | 2480332 |
| Chordata | Aves | Charadriiformes | Charadriidae | Pluvialis | Pluvialis squatarola | 2480327 |
| Chordata | Aves | Charadriiformes | Charadriidae | Charadrius | Charadrius hiaticula | 9566659 |
| Chordata | Aves | Charadriiformes | Charadriidae | Charadrius | Charadrius dubius | 7937336 |
| Chordata | Aves | Procellariiformes | Procellariidae | Puffinus | Puffinus mauretanicus | 5229360 |
| Chordata | Aves | Charadriiformes | Charadriidae | Charadrius | Charadrius alexandrinus | 2480311 |
| Chordata | Aves | Charadriiformes | Charadriidae | Charadrius | Charadrius leschenaultii | 2480297 |
| Chordata | Aves | Charadriiformes | Charadriidae | Charadrius | Charadrius asiaticus | 2480308 |
| Chordata | Aves | Charadriiformes | Charadriidae | Charadrius | Charadrius morinellus | 2480281 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Scolopax | Scolopax rusticola | 2481700 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Lymnocryptes | Lymnocryptes minimus | 2481835 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Gallinago | Gallinago stenura | 2481828 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Gallinago | Gallinago media | 2481831 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Gallinago | Gallinago gallinago | 2481819 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Limosa | Limosa limosa | 2481685 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Limosa | Limosa lapponica | 2481681 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Numenius | Numenius phaeopus | 2481784 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Numenius | Numenius arquata | 2481792 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Tringa | Tringa erythropus | 2481725 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Tringa | Tringa totanus | 2481714 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Tringa | Tringa stagnatilis | 2481719 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Tringa | Tringa nebularia | 2481726 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Tringa | Tringa ochropus | 2481728 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Tringa | Tringa glareola | 2481713 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Xenus | Xenus cinereus | 2481703 |
| Chordata | Aves | Procellariiformes | Hydrobatidae | Pelagodroma | Pelagodroma marina | 2481991 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Actitis | Actitis hypoleucos | 2481800 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Arenaria | Arenaria interpres | 2481776 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Calidris | Calidris minuta | 2481749 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Calidris | Calidris temminckii | 2481740 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Calidris | Calidris alpina | 2481759 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Calidris | Calidris maritima | 2481743 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Calidris | Calidris falcinellus | 7537888 |
| Chordata | Aves | Gaviiformes | Gaviidae | Gavia | Gavia arctica | 2481959 |
| Chordata | Aves | Procellariiformes | Hydrobatidae | Hydrobates | Hydrobates pelagicus | 4408455 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Calidris | Calidris pugnax | 8250742 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Phalaropus | Phalaropus lobatus | 5739290 |
| Chordata | Aves | Charadriiformes | Scolopacidae | Phalaropus | Phalaropus fulicarius | 5229390 |
| Chordata | Aves | Charadriiformes | Stercorariidae | Stercorarius | Stercorarius skua | 2481618 |
| Chordata | Aves | Charadriiformes | Stercorariidae | Stercorarius | Stercorarius pomarinus | 2481609 |
| Chordata | Aves | Charadriiformes | Stercorariidae | Stercorarius | Stercorarius parasiticus | 2481621 |
| Chordata | Aves | Charadriiformes | Stercorariidae | Stercorarius | Stercorarius longicaudus | 2481613 |
| Chordata | Aves | Charadriiformes | Laridae | Larus | Larus canus | 7982160 |
| Chordata | Aves | Charadriiformes | Laridae | Ichthyaetus | Ichthyaetus audouinii | 7406504 |
| Chordata | Aves | Charadriiformes | Laridae | Larus | Larus marinus | 2481172 |
| Chordata | Aves | Procellariiformes | Hydrobatidae | Oceanodroma | Oceanodroma leucorhoa | 2482012 |
| Chordata | Aves | Charadriiformes | Laridae | Larus | Larus hyperboreus | 2481127 |
| Chordata | Aves | Charadriiformes | Laridae | Larus | Larus argentatus | 2481139 |
| Chordata | Aves | Charadriiformes | Laridae | Larus | Larus fuscus | 2481174 |
| Chordata | Aves | Charadriiformes | Laridae | Larus | Larus cachinnans | 2481181 |
| Chordata | Aves | Charadriiformes | Laridae | Larus | Larus armenicus | 2481171 |
| Chordata | Aves | Charadriiformes | Laridae | Ichthyaetus | Ichthyaetus ichthyaetus | 8028588 |
| Chordata | Aves | Charadriiformes | Laridae | Chroicocephalus | Chroicocephalus ridibundus | 6065824 |
| Chordata | Aves | Charadriiformes | Laridae | Chroicocephalus | Chroicocephalus genei | 6065810 |
| Chordata | Aves | Charadriiformes | Laridae | Ichthyaetus | Ichthyaetus melanocephalus | 8151548 |
| Chordata | Aves | Procellariiformes | Hydrobatidae | Oceanodroma | Oceanodroma castro | 2482001 |
| Chordata | Aves | Charadriiformes | Laridae | Hydrocoloeus | Hydrocoloeus minutus | 6065841 |
| Chordata | Aves | Charadriiformes | Laridae | Pagophila | Pagophila eburnea | 2481202 |
| Chordata | Aves | Charadriiformes | Laridae | Rissa | Rissa tridactyla | 2481205 |
| Chordata | Aves | Charadriiformes | Laridae | Gelochelidon | Gelochelidon nilotica | 2481263 |
| Chordata | Aves | Charadriiformes | Laridae | Hydroprogne | Hydroprogne caspia | 2481236 |
| Chordata | Aves | Charadriiformes | Laridae | Thalasseus | Thalasseus bengalensis | 6065833 |
| Chordata | Aves | Charadriiformes | Laridae | Thalasseus | Thalasseus sandvicensis | 2481240 |
| Chordata | Aves | Charadriiformes | Laridae | Sterna | Sterna dougallii | 5229249 |
| Chordata | Aves | Charadriiformes | Laridae | Sterna | Sterna hirundo | 9367409 |
| Chordata | Aves | Phaethontiformes | Phaethontidae | Phaethon | Phaethon aethereus | 9743996 |
| Chordata | Aves | Charadriiformes | Laridae | Sterna | Sterna paradisaea | 5229230 |
| Chordata | Aves | Charadriiformes | Laridae | Sternula | Sternula albifrons | 5789279 |
| Chordata | Aves | Charadriiformes | Laridae | Chlidonias | Chlidonias hybrida | 2481121 |
| Chordata | Aves | Charadriiformes | Laridae | Chlidonias | Chlidonias leucopterus | 2481120 |
| Chordata | Aves | Charadriiformes | Laridae | Chlidonias | Chlidonias niger | 2481119 |
| Chordata | Aves | Charadriiformes | Alcidae | Alle | Alle alle | 2481299 |
| Chordata | Aves | Charadriiformes | Alcidae | Uria | Uria aalge | 2481342 |
| Chordata | Aves | Charadriiformes | Alcidae | Uria | Uria lomvia | 2481339 |
| Chordata | Aves | Charadriiformes | Alcidae | Alca | Alca torda | 8277073 |
| Chordata | Aves | Suliformes | Sulidae | Morus | Morus bassanus | 2480964 |
| Chordata | Aves | Charadriiformes | Alcidae | Cepphus | Cepphus grylle | 2481312 |
| Chordata | Aves | Charadriiformes | Alcidae | Fratercula | Fratercula arctica | 2481353 |
| Chordata | Aves | Pteroclidiformes | Pteroclididae | Pterocles | Pterocles alchata | 2480204 |
| Chordata | Aves | Pteroclidiformes | Pteroclididae | Pterocles | Pterocles orientalis | 2480193 |
| Chordata | Aves | Columbiformes | Columbidae | Columba | Columba livia | 2495414 |
| Chordata | Aves | Columbiformes | Columbidae | Columba | Columba oenas | 2495471 |
| Chordata | Aves | Columbiformes | Columbidae | Columba | Columba palumbus | 2495455 |
| Chordata | Aves | Columbiformes | Columbidae | Columba | Columba trocaz | 2495406 |
| Chordata | Aves | Columbiformes | Columbidae | Streptopelia | Streptopelia turtur | 2495708 |
| Chordata | Aves | Columbiformes | Columbidae | Streptopelia | Streptopelia decaocto | 2495696 |
| Chordata | Aves | Suliformes | Phalacrocoracidae | Phalacrocorax | Phalacrocorax carbo | 2481890 |
| Chordata | Aves | Columbiformes | Columbidae | Spilopelia | Streptopelia senegalensis | 6101223 |
| Chordata | Aves | Cuculiformes | Cuculidae | Clamator | Clamator glandarius | 2496464 |
| Chordata | Aves | Cuculiformes | Cuculidae | Cuculus | Cuculus canorus | 5231918 |
| Chordata | Aves | Cuculiformes | Cuculidae | Cuculus | Cuculus saturatus | 9548093 |
| Chordata | Aves | Strigiformes | Tytonidae | Tyto | Tyto alba | 2497921 |
| Chordata | Aves | Strigiformes | Strigidae | Otus | Otus brucei | 2497755 |
| Chordata | Aves | Strigiformes | Strigidae | Otus | Otus scops | 2497701 |
| Chordata | Aves | Strigiformes | Strigidae | Bubo | Bubo bubo | 5959092 |
| Chordata | Aves | Strigiformes | Strigidae | Bubo | Bubo scandiacus | 5959143 |
| Chordata | Aves | Strigiformes | Strigidae | Strix | Strix aluco | 9282206 |
| Chordata | Aves | Suliformes | Phalacrocoracidae | Phalacrocorax | Phalacrocorax aristotelis | 2481861 |
| Chordata | Aves | Strigiformes | Strigidae | Strix | Strix uralensis | 2497509 |
| Chordata | Aves | Strigiformes | Strigidae | Strix | Strix nebulosa | 2497522 |
| Chordata | Aves | Strigiformes | Strigidae | Surnia | Surnia ulula | 2497894 |
| Chordata | Aves | Strigiformes | Strigidae | Glaucidium | Glaucidium passerinum | 5232162 |
| Chordata | Aves | Strigiformes | Strigidae | Athene | Athene noctua | 2497266 |
| Chordata | Aves | Strigiformes | Strigidae | Aegolius | Aegolius funereus | 5739298 |
| Chordata | Aves | Strigiformes | Strigidae | Asio | Asio otus | 2497306 |
| Chordata | Aves | Strigiformes | Strigidae | Asio | Asio flammeus | 2497295 |
| Chordata | Aves | Caprimulgiformes | Caprimulgidae | Caprimulgus | Caprimulgus ruficollis | 9411857 |
| Chordata | Aves | Caprimulgiformes | Caprimulgidae | Caprimulgus | Caprimulgus europaeus | 8109681 |
| Chordata | Aves | Suliformes | Phalacrocoracidae | Microcarbo | Microcarbo pygmaeus | 10095118 |
| Chordata | Aves | Apodiformes | Apodidae | Tachymarptis | Apus melba | 2477411 |
| Chordata | Aves | Apodiformes | Apodidae | Apus | Apus apus | 5228676 |
| Chordata | Aves | Apodiformes | Apodidae | Apus | Apus unicolor | 5228694 |
| Chordata | Aves | Apodiformes | Apodidae | Apus | Apus pallidus | 5228695 |
| Chordata | Aves | Apodiformes | Apodidae | Apus | Apus affinis | 5228662 |
| Chordata | Aves | Apodiformes | Apodidae | Apus | Apus caffer | 5228644 |
| Chordata | Aves | Coraciiformes | Alcedinidae | Alcedo | Alcedo atthis | 2475532 |
| Chordata | Aves | Coraciiformes | Alcedinidae | Halcyon | Halcyon smyrnensis | 5228328 |
| Chordata | Aves | Coraciiformes | Alcedinidae | Ceryle | Ceryle rudis | 2475679 |
| Chordata | Aves | Pelecaniformes | Pelecanidae | Pelecanus | Pelecanus onocrotalus | 2480350 |
| Chordata | Aves | Coraciiformes | Meropidae | Merops | Merops apiaster | 2475443 |
| Chordata | Aves | Coraciiformes | Coraciidae | Coracias | Coracias garrulus | 2475365 |
| Chordata | Aves | Bucerotiformes | Upupidae | Upupa | Upupa epops | 2498415 |
| Chordata | Aves | Piciformes | Picidae | Jynx | Jynx torquilla | 8012314 |
| Chordata | Aves | Piciformes | Picidae | Dryobates | Dryobates minor | 9015784 |
| Chordata | Aves | Piciformes | Picidae | Dendrocoptes | Dendrocoptes medius | 8773033 |
| Chordata | Aves | Piciformes | Picidae | Dendrocopos | Dendrocopos major | 2477968 |
| Chordata | Aves | Piciformes | Picidae | Dendrocopos | Dendrocopos syriacus | 2478031 |
| Chordata | Aves | Piciformes | Picidae | Picoides | Picoides tridactylus | 2477804 |
| Chordata | Aves | Pelecaniformes | Pelecanidae | Pelecanus | Pelecanus crispus | 2480353 |
| Chordata | Aves | Piciformes | Picidae | Dryocopus | Dryocopus martius | 2477872 |
| Chordata | Aves | Piciformes | Picidae | Picus | Picus viridis | 2478523 |
| Chordata | Aves | Piciformes | Picidae | Picus | Picus canus | 2478548 |
| Chordata | Aves | Passeriformes | Alaudidae | Ammomanes | Ammomanes deserti | 2490641 |
| Chordata | Aves | Passeriformes | Alaudidae | Melanocorypha | Melanocorypha calandra | 2490613 |
| Chordata | Aves | Passeriformes | Alaudidae | Melanocorypha | Melanocorypha bimaculata | 2490609 |
| Chordata | Aves | Passeriformes | Alaudidae | Melanocorypha | Alauda leucoptera | 2490611 |
| Chordata | Aves | Passeriformes | Alaudidae | Calandrella | Calandrella brachydactyla | 2490681 |
| Chordata | Aves | Passeriformes | Alaudidae | Calandrella | Alaudala rufescens | 2490682 |
| Chordata | Aves | Gaviiformes | Gaviidae | Gavia | Gavia immer | 2481962 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Ardea | Ardea cinerea | 9797180 |
| Chordata | Aves | Passeriformes | Alaudidae | Chersophilus | Chersophilus duponti | 2490674 |
| Chordata | Aves | Passeriformes | Alaudidae | Galerida | Galerida cristata | 2490669 |
| Chordata | Aves | Passeriformes | Alaudidae | Lullula | Lullula arborea | 2490604 |
| Chordata | Aves | Passeriformes | Alaudidae | Alauda | Alauda arvensis | 8077224 |
| Chordata | Aves | Passeriformes | Alaudidae | Eremophila | Eremophila alpestris | 9415596 |
| Chordata | Aves | Passeriformes | Hirundinidae | Riparia | Riparia riparia | 2489160 |
| Chordata | Aves | Passeriformes | Hirundinidae | Ptyonoprogne | Ptyonoprogne rupestris | 4408796 |
| Chordata | Aves | Passeriformes | Hirundinidae | Hirundo | Hirundo rustica | 9515886 |
| Chordata | Aves | Passeriformes | Hirundinidae | Cecropis | Cecropis daurica | 9717283 |
| Chordata | Aves | Passeriformes | Hirundinidae | Delichon | Delichon urbicum | 2489214 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Ardea | Ardea purpurea | 2480934 |
| Chordata | Aves | Passeriformes | Motacillidae | Motacilla | Motacilla alba | 9687165 |
| Chordata | Aves | Passeriformes | Motacillidae | Motacilla | Motacilla citreola | 2490307 |
| Chordata | Aves | Passeriformes | Motacillidae | Motacilla | Motacilla flava | 9701857 |
| Chordata | Aves | Passeriformes | Motacillidae | Motacilla | Motacilla cinerea | 2490310 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus campestris | 2490274 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus berthelotii | 2490270 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus trivialis | 2490246 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus hodgsoni | 2490244 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus pratensis | 2490266 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus cervinus | 2490283 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Ardea | Ardea alba | 9752617 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus petrosus | 2490275 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus spinoletta | 2490255 |
| Chordata | Aves | Passeriformes | Pycnonotidae | Pycnonotus | Pycnonotus xanthopygos | 2486116 |
| Chordata | Aves | Passeriformes | Regulidae | Regulus | Regulus regulus | 2484596 |
| Chordata | Aves | Passeriformes | Regulidae | Regulus | Regulus ignicapilla | 6093527 |
| Chordata | Aves | Passeriformes | Bombycillidae | Bombycilla | Bombycilla garrulus | 2484605 |
| Chordata | Aves | Passeriformes | Cinclidae | Cinclus | Cinclus cinclus | 2495093 |
| Chordata | Aves | Passeriformes | Troglodytidae | Troglodytes | Troglodytes troglodytes | 5231438 |
| Chordata | Aves | Passeriformes | Prunellidae | Prunella | Prunella collaris | 5231770 |
| Chordata | Aves | Passeriformes | Prunellidae | Prunella | Prunella montanella | 5231759 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Egretta | Egretta garzetta | 2480876 |
| Chordata | Aves | Passeriformes | Prunellidae | Prunella | Prunella ocularis | 5231773 |
| Chordata | Aves | Passeriformes | Prunellidae | Prunella | Prunella atrogularis | 5231766 |
| Chordata | Aves | Passeriformes | Prunellidae | Prunella | Prunella modularis | 5231763 |
| Chordata | Aves | Passeriformes | Muscicapidae | Monticola | Monticola saxatilis | 2490956 |
| Chordata | Aves | Passeriformes | Muscicapidae | Monticola | Monticola solitarius | 2490955 |
| Chordata | Aves | Passeriformes | Turdidae | Zoothera | Zoothera aurea | 4408785 |
| Chordata | Aves | Passeriformes | Turdidae | Turdus | Turdus torquatus | 7865305 |
| Chordata | Aves | Passeriformes | Turdidae | Turdus | Turdus merula | 2490719 |
| Chordata | Aves | Passeriformes | Turdidae | Turdus | Turdus pilaris | 2490737 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Ardeola | Ardeola ralloides | 2480906 |
| Chordata | Aves | Passeriformes | Turdidae | Turdus | Turdus iliacus | 2490781 |
| Chordata | Aves | Passeriformes | Turdidae | Turdus | Turdus philomelos | 7901064 |
| Chordata | Aves | Passeriformes | Turdidae | Turdus | Turdus viscivorus | 2490774 |
| Chordata | Aves | Passeriformes | Cisticolidae | Cisticola | Cisticola juncidis | 2492822 |
| Chordata | Aves | Passeriformes | Cisticolidae | Prinia | Prinia gracilis | 2492748 |
| Chordata | Aves | Passeriformes | Cettiidae | Cettia | Cettia cetti | 2493012 |
| Chordata | Aves | Passeriformes | Locustellidae | Locustella | Locustella lanceolata | 2493550 |
| Chordata | Aves | Passeriformes | Locustellidae | Locustella | Locustella naevia | 2493548 |
| Chordata | Aves | Passeriformes | Locustellidae | Locustella | Locustella fluviatilis | 2493549 |
| Chordata | Aves | Passeriformes | Locustellidae | Locustella | Locustella luscinioides | 2493551 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Bubulcus | Bubulcus ibis | 2480830 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Acrocephalus | Acrocephalus melanopogon | 2493124 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Acrocephalus | Acrocephalus paludicola | 2493114 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Acrocephalus | Acrocephalus schoenobaenus | 2493129 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Acrocephalus | Acrocephalus agricola | 2493137 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Acrocephalus | Acrocephalus scirpaceus | 2493118 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Acrocephalus | Acrocephalus dumetorum | 2493145 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Acrocephalus | Acrocephalus palustris | 2493136 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Acrocephalus | Acrocephalus arundinaceus | 2493128 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Iduna | Iduna caligata | 5739359 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Iduna | Iduna pallida | 5739361 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Nycticorax | Nycticorax nycticorax | 2480863 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Hippolais | Hippolais languida | 2493217 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Hippolais | Hippolais olivetorum | 2493219 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Hippolais | Hippolais polyglotta | 2493214 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Hippolais | Hippolais icterina | 2493220 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus trochilus | 2493052 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus collybita | 2493091 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus bonelli | 2493070 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus sibilatrix | 8128385 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus inornatus | 2493095 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Ixobrychus | Ixobrychus minutus | 2480850 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus borealis | 2493071 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus trochiloides | 2493084 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia atricapilla | 2492956 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia borin | 2492942 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia communis | 2492943 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia curruca | 2492960 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia nisoria | 2492955 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia hortensis | 2492944 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia ruppeli | 6100917 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia cantillans | 2492950 |
| Chordata | Aves | Pelecaniformes | Ardeidae | Botaurus | Botaurus stellaris | 2480910 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia melanocephala | 2492962 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia melanothorax | 2492953 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia mystacea | 2492954 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia conspicillata | 2492959 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia undata | 2492949 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia sarda | 2492965 |
| Chordata | Aves | Passeriformes | Muscicapidae | Muscicapa | Muscicapa striata | 2492576 |
| Chordata | Aves | Passeriformes | Muscicapidae | Ficedula | Ficedula hypoleuca | 2492606 |
| Chordata | Aves | Passeriformes | Muscicapidae | Ficedula | Ficedula albicollis | 2492601 |
| Chordata | Aves | Passeriformes | Muscicapidae | Ficedula | Ficedula semitorquata | 2492608 |
| Chordata | Aves | Ciconiiformes | Ciconiidae | Ciconia | Ciconia nigra | 2481909 |
| Chordata | Aves | Passeriformes | Muscicapidae | Ficedula | Ficedula parva | 2492605 |
| Chordata | Aves | Passeriformes | Muscicapidae | Erithacus | Erithacus rubecula | 2492462 |
| Chordata | Aves | Passeriformes | Muscicapidae | Luscinia | Luscinia luscinia | 2492542 |
| Chordata | Aves | Passeriformes | Muscicapidae | Luscinia | Luscinia megarhynchos | 7660710 |
| Chordata | Aves | Passeriformes | Muscicapidae | Luscinia | Calliope calliope | 2492537 |
| Chordata | Aves | Passeriformes | Muscicapidae | Luscinia | Luscinia svecica | 4408732 |
| Chordata | Aves | Passeriformes | Muscicapidae | Tarsiger | Tarsiger cyanurus | 2492509 |
| Chordata | Aves | Passeriformes | Muscicapidae | Irania | Irania gutturalis | 2492700 |
| Chordata | Aves | Passeriformes | Muscicapidae | Erythropygia | Cercotrichas galactotes | 5788860 |
| Chordata | Aves | Passeriformes | Muscicapidae | Phoenicurus | Phoenicurus ochruros | 5739315 |
| Chordata | Aves | Gaviiformes | Gaviidae | Gavia | Gavia adamsii | 2481957 |
| Chordata | Aves | Ciconiiformes | Ciconiidae | Ciconia | Ciconia ciconia | 2481912 |
| Chordata | Aves | Passeriformes | Muscicapidae | Phoenicurus | Phoenicurus phoenicurus | 5739317 |
| Chordata | Aves | Passeriformes | Muscicapidae | Saxicola | Saxicola rubetra | 2492521 |
| Chordata | Aves | Passeriformes | Muscicapidae | Saxicola | Saxicola rubicola | 4408759 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe leucura | 5231236 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe oenanthe | 5231240 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe finschii | 5231251 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe pleschanka | 5231233 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe hispanica | 5231235 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe chrysopygia | 5788868 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe deserti | 5231244 |
| Chordata | Aves | Pelecaniformes | Threskiornithidae | Plegadis | Plegadis falcinellus | 2480773 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe cypriaca | 5231234 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe isabellina | 5231245 |
| Chordata | Aves | Passeriformes | Panuridae | Panurus | Panurus biarmicus | 2493367 |
| Chordata | Aves | Passeriformes | Aegithalidae | Aegithalos | Aegithalos caudatus | 2495000 |
| Chordata | Aves | Passeriformes | Paridae | Poecile | Poecile lugubris | 2487857 |
| Chordata | Aves | Passeriformes | Paridae | Poecile | Poecile palustris | 2487843 |
| Chordata | Aves | Passeriformes | Paridae | Poecile | Poecile montanus | 2487866 |
| Chordata | Aves | Passeriformes | Paridae | Poecile | Poecile cinctus | 2487862 |
| Chordata | Aves | Passeriformes | Paridae | Periparus | Periparus ater | 2487871 |
| Chordata | Aves | Passeriformes | Paridae | Lophophanes | Lophophanes cristatus | 2487883 |
| Chordata | Aves | Pelecaniformes | Threskiornithidae | Platalea | Platalea leucorodia | 2480801 |
| Chordata | Aves | Passeriformes | Paridae | Parus | Parus major | 9705453 |
| Chordata | Aves | Passeriformes | Paridae | Cyanistes | Cyanistes caeruleus | 2487879 |
| Chordata | Aves | Passeriformes | Paridae | Cyanistes | Cyanistes cyanus | 2487877 |
| Chordata | Aves | Passeriformes | Sittidae | Sitta | Sitta europaea | 2484916 |
| Chordata | Aves | Passeriformes | Sittidae | Sitta | Sitta whiteheadi | 2484886 |
| Chordata | Aves | Passeriformes | Sittidae | Sitta | Sitta krueperi | 2484880 |
| Chordata | Aves | Passeriformes | Sittidae | Sitta | Sitta neumayer | 2484890 |
| Chordata | Aves | Passeriformes | Sittidae | Sitta | Sitta tephronota | 2484885 |
| Chordata | Aves | Passeriformes | Tichodromidae | Tichodroma | Tichodroma muraria | 2484918 |
| Chordata | Aves | Passeriformes | Certhiidae | Certhia | Certhia familiaris | 2487603 |
| Chordata | Aves | Phoenicopteriformes | Phoenicopteridae | Phoenicopterus | Phoenicopterus roseus | 4352332 |
| Chordata | Aves | Passeriformes | Certhiidae | Certhia | Certhia brachydactyla | 7806848 |
| Chordata | Aves | Passeriformes | Remizidae | Remiz | Remiz pendulinus | 2487385 |
| Chordata | Aves | Passeriformes | Oriolidae | Oriolus | Oriolus oriolus | 2488949 |
| Chordata | Aves | Passeriformes | Laniidae | Lanius | Lanius collurio | 7745240 |
| Chordata | Aves | Passeriformes | Laniidae | Lanius | Lanius excubitor | 9444556 |
| Chordata | Aves | Passeriformes | Laniidae | Lanius | Lanius meridionalis | 7341500 |
| Chordata | Aves | Passeriformes | Laniidae | Lanius | Lanius minor | 2492854 |
| Chordata | Aves | Passeriformes | Laniidae | Lanius | Lanius nubicus | 2492855 |
| Chordata | Aves | Passeriformes | Laniidae | Lanius | Lanius senator | 2492867 |
| Chordata | Aves | Passeriformes | Corvidae | Perisoreus | Perisoreus infaustus | 2482449 |
| Chordata | Aves | Anseriformes | Anatidae | Cygnus | Cygnus olor | 2498343 |
| Chordata | Aves | Passeriformes | Corvidae | Garrulus | Garrulus glandarius | 5229493 |
| Chordata | Aves | Passeriformes | Corvidae | Pica | Pica pica | 5229490 |
| Chordata | Aves | Passeriformes | Corvidae | Nucifraga | Nucifraga caryocatactes | 2482613 |
| Chordata | Aves | Passeriformes | Corvidae | Pyrrhocorax | Pyrrhocorax pyrrhocorax | 2482552 |
| Chordata | Aves | Passeriformes | Corvidae | Pyrrhocorax | Pyrrhocorax graculus | 2482553 |
| Chordata | Aves | Passeriformes | Corvidae | Coloeus | Corvus monedula | 6100954 |
| Chordata | Aves | Passeriformes | Corvidae | Corvus | Corvus frugilegus | 2482513 |
| Chordata | Aves | Passeriformes | Corvidae | Corvus | Corvus corone | 9409796 |
| Chordata | Aves | Passeriformes | Corvidae | Corvus | Corvus corax | 2482492 |
| Chordata | Aves | Anseriformes | Anatidae | Cygnus | Cygnus cygnus | 2498347 |
| Chordata | Aves | Passeriformes | Sturnidae | Pastor | Pastor roseus | 5845386 |
| Chordata | Aves | Passeriformes | Sturnidae | Sturnus | Sturnus vulgaris | 9809229 |
| Chordata | Aves | Passeriformes | Sturnidae | Sturnus | Sturnus unicolor | 2489104 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza citrinella | 2491534 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza leucocephalos | 2491472 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza cirlus | 7626513 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza cia | 2491498 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza buchanani | 9566476 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza cineracea | 2491469 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza hortulana | 2491482 |
| Chordata | Aves | Anseriformes | Anatidae | Cygnus | Cygnus columbianus | 2498338 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza caesia | 2491490 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza pusilla | 2491544 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza rustica | 2491582 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza aureola | 2491518 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza melanocephala | 9613112 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza bruniceps | 2491521 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza pallasi | 2491591 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza schoeniclus | 2491557 |
| Chordata | Aves | Passeriformes | Emberizidae | Emberiza | Emberiza calandra | 7634625 |
| Chordata | Aves | Passeriformes | Calcariidae | Calcarius | Calcarius lapponicus | 2491678 |
| Chordata | Aves | Anseriformes | Anatidae | Anser | Anser fabalis | 2498027 |
| Chordata | Aves | Passeriformes | Calcariidae | Plectrophenax | Plectrophenax nivalis | 2491719 |
| Chordata | Aves | Passeriformes | Fringillidae | Fringilla | Fringilla coelebs | 2494422 |
| Chordata | Aves | Passeriformes | Fringillidae | Fringilla | Fringilla montifringilla | 2494441 |
| Chordata | Aves | Passeriformes | Fringillidae | Pinicola | Pinicola enucleator | 2494771 |
| Chordata | Aves | Passeriformes | Fringillidae | Carpodacus | Carpodacus erythrinus | 2494459 |
| Chordata | Aves | Passeriformes | Fringillidae | Carpodacus | Carpodacus rubicilla | 2494517 |
| Chordata | Aves | Passeriformes | Fringillidae | Loxia | Loxia pytyopsittacus | 2494154 |
| Chordata | Aves | Passeriformes | Fringillidae | Loxia | Loxia scotica | 2494177 |
| Chordata | Aves | Passeriformes | Fringillidae | Loxia | Loxia curvirostra | 9629160 |
| Chordata | Aves | Passeriformes | Fringillidae | Loxia | Loxia leucoptera | 8144830 |
| Chordata | Aves | Anseriformes | Anatidae | Anser | Anser brachyrhynchus | 2498024 |
| Chloris chloris | 0 | |||||
| Chordata | Aves | Passeriformes | Fringillidae | Acanthis | Acanthis flammea | 5231630 |
| Chordata | Aves | Passeriformes | Fringillidae | Acanthis | Acanthis hornemanni | 5231646 |
| Chordata | Aves | Passeriformes | Fringillidae | Spinus | Spinus spinus | 6092830 |
| Chordata | Aves | Passeriformes | Fringillidae | Carduelis | Carduelis carduelis | 2494686 |
| Chordata | Aves | Passeriformes | Fringillidae | Linaria | Linaria flavirostris | 7668711 |
| Chordata | Aves | Passeriformes | Fringillidae | Linaria | Linaria cannabina | 8104397 |
| Chordata | Aves | Passeriformes | Fringillidae | Serinus | Serinus pusillus | 2494194 |
| Chordata | Aves | Passeriformes | Fringillidae | Serinus | Serinus serinus | 2494200 |
| Chordata | Aves | Passeriformes | Fringillidae | Serinus | Serinus canaria | 2494303 |
| Chordata | Aves | Anseriformes | Anatidae | Anser | Anser albifrons | 2498017 |
| Chordata | Aves | Passeriformes | Fringillidae | Carduelis | Carduelis citrinella | 2494632 |
| Chordata | Aves | Passeriformes | Fringillidae | Pyrrhula | Pyrrhula pyrrhula | 2494543 |
| Chordata | Aves | Passeriformes | Fringillidae | Pyrrhula | Pyrrhula murina | 4408970 |
| Chordata | Aves | Passeriformes | Fringillidae | Coccothraustes | Coccothraustes coccothraustes | 2494329 |
| Chordata | Aves | Passeriformes | Fringillidae | Rhodopechys | Rhodopechys sanguineus | 2494570 |
| Chordata | Aves | Passeriformes | Fringillidae | Bucanetes | Bucanetes githagineus | 2494394 |
| Chordata | Aves | Passeriformes | Fringillidae | Bucanetes | Bucanetes mongolicus | 4408954 |
| Chordata | Aves | Passeriformes | Fringillidae | Rhodospiza | Rhodospiza obsoleta | 2494787 |
| Chordata | Aves | Passeriformes | Passeridae | Passer | Passer italiae | 8426551 |
| Chordata | Aves | Passeriformes | Passeridae | Passer | Passer domesticus | 5231190 |
| Chordata | Aves | Podicipediformes | Podicipedidae | Tachybaptus | Tachybaptus ruficollis | 2482048 |
| Chordata | Aves | Anseriformes | Anatidae | Anser | Anser erythropus | 2498026 |
| Chordata | Aves | Passeriformes | Passeridae | Passer | Passer hispaniolensis | 5231209 |
| Chordata | Aves | Passeriformes | Passeridae | Passer | Passer moabiticus | 5231203 |
| Chordata | Aves | Passeriformes | Passeridae | Passer | Passer montanus | 5231198 |
| Chordata | Aves | Passeriformes | Passeridae | Gymnoris | Gymnoris xanthocollis | 6100968 |
| Chordata | Aves | Passeriformes | Passeridae | Petronia | Petronia petronia | 2492350 |
| Chordata | Aves | Passeriformes | Passeridae | Carpospiza | Carpospiza brachydactyla | 2492364 |
| Chordata | Aves | Passeriformes | Passeridae | Montifringilla | Montifringilla nivalis | 2492371 |
| Chordata | Aves | Passeriformes | Alaudidae | Galerida | Galerida theklae | 9330514 |
| Chordata | Aves | Columbiformes | Columbidae | Columba | Columba bollii | 2495427 |
| Chordata | Aves | Columbiformes | Columbidae | Columba | Columba junoniae | 2495434 |
| Chordata | Aves | Anseriformes | Anatidae | Anser | Anser anser | 2498036 |
| Chordata | Aves | Passeriformes | Fringillidae | Fringilla | Fringilla teydea | 2494442 |
| Chordata | Aves | Passeriformes | Muscicapidae | Saxicola | Saxicola dacotiae | 2492522 |
| Chordata | Aves | Charadriiformes | Charadriidae | Vanellus | Vanellus gregarius | 5229138 |
| Chordata | Aves | Otidiformes | Otididae | Chlamydotis | Chlamydotis undulata | 2474864 |
| Chordata | Aves | Charadriiformes | Glareolidae | Cursorius | Cursorius cursor | 2480740 |
| Chordata | Aves | Anseriformes | Anatidae | Branta | Branta leucopsis | 5232464 |
| Chordata | Aves | Anseriformes | Anatidae | Tadorna | Tadorna ferruginea | 2498015 |
| Chordata | Aves | Anseriformes | Anatidae | Tadorna | Tadorna tadorna | 2498009 |
| Chordata | Aves | Anseriformes | Anatidae | Mareca | Mareca penelope | 8000602 |
| Chordata | Aves | Anseriformes | Anatidae | Mareca | Mareca strepera | 9362027 |
| Chordata | Aves | Podicipediformes | Podicipedidae | Podiceps | Podiceps cristatus | 2482054 |
| Chordata | Aves | Anseriformes | Anatidae | Anas | Anas crecca | 8214667 |
| Chordata | Aves | Anseriformes | Anatidae | Anas | Anas platyrhynchos | 9761484 |
| Chordata | Aves | Anseriformes | Anatidae | Anas | Anas acuta | 2498112 |
| Chordata | Aves | Anseriformes | Anatidae | Spatula | Spatula querquedula | 9274012 |
| Chordata | Aves | Anseriformes | Anatidae | Spatula | Spatula clypeata | 8332393 |
| Chordata | Aves | Anseriformes | Anatidae | Marmaronetta | Marmaronetta angustirostris | 2498390 |
| Chordata | Aves | Anseriformes | Anatidae | Netta | Netta rufina | 2498180 |
| Chordata | Aves | Anseriformes | Anatidae | Aythya | Aythya ferina | 2498255 |
| Chordata | Aves | Anseriformes | Anatidae | Aythya | Aythya nyroca | 2498259 |
| Chordata | Aves | Anseriformes | Anatidae | Aythya | Aythya fuligula | 2498261 |
| Chordata | Aves | Podicipediformes | Podicipedidae | Podiceps | Podiceps grisegena | 2482051 |
| Chordata | Aves | Anseriformes | Anatidae | Aythya | Aythya marila | 2498265 |
| Chordata | Aves | Anseriformes | Anatidae | Somateria | Somateria mollissima | 2498352 |
| Chordata | Aves | Anseriformes | Anatidae | Somateria | Somateria spectabilis | 2498350 |
| Chordata | Aves | Anseriformes | Anatidae | Polysticta | Polysticta stelleri | 5232431 |
| Chordata | Aves | Anseriformes | Anatidae | Histrionicus | Histrionicus histrionicus | 2498224 |
| Chordata | Aves | Anseriformes | Anatidae | Clangula | Clangula hyemalis | 2498273 |
| Chordata | Aves | Anseriformes | Anatidae | Melanitta | Melanitta nigra | 2498247 |
| Chordata | Aves | Anseriformes | Anatidae | Melanitta | Melanitta fusca | 2498238 |
| Chordata | Aves | Anseriformes | Anatidae | Bucephala | Bucephala clangula | 2498326 |
| Chordata | Aves | Anseriformes | Anatidae | Bucephala | Bucephala islandica | 2498325 |
| Chordata | Aves | Podicipediformes | Podicipedidae | Podiceps | Podiceps auritus | 2482059 |
| Chordata | Aves | Anseriformes | Anatidae | Mergellus | Mergellus albellus | 2498285 |
| Chordata | Aves | Anseriformes | Anatidae | Mergus | Mergus serrator | 2498375 |
| Chordata | Aves | Anseriformes | Anatidae | Mergus | Mergus merganser | 9537647 |
| Chordata | Aves | Anseriformes | Anatidae | Oxyura | Oxyura leucocephala | 2498302 |
| Chordata | Aves | Accipitriformes | Pandionidae | Pandion | Pandion haliaetus | 2480726 |
| Chordata | Aves | Accipitriformes | Accipitridae | Pernis | Pernis apivorus | 2480420 |
| Chordata | Aves | Accipitriformes | Accipitridae | Elanus | Elanus caeruleus | 2480372 |
| Chordata | Aves | Accipitriformes | Accipitridae | Milvus | Milvus milvus | 5229168 |
| Chordata | Aves | Accipitriformes | Accipitridae | Milvus | Milvus migrans | 5229167 |
| Chordata | Aves | Podicipediformes | Podicipedidae | Podiceps | Podiceps nigricollis | 2482065 |
| Chordata | Aves | Accipitriformes | Accipitridae | Haliaeetus | Haliaeetus albicilla | 2480449 |
| Chordata | Aves | Accipitriformes | Accipitridae | Gypaetus | Gypaetus barbatus | 2480649 |
| Chordata | Aves | Accipitriformes | Accipitridae | Neophron | Neophron percnopterus | 2480696 |
| Chordata | Aves | Accipitriformes | Accipitridae | Gyps | Gyps fulvus | 2480389 |
| Chordata | Aves | Accipitriformes | Accipitridae | Aegypius | Aegypius monachus | 5229165 |
| Chordata | Aves | Accipitriformes | Accipitridae | Circaetus | Circaetus gallicus | 2480666 |
| Chordata | Aves | Accipitriformes | Accipitridae | Circus | Circus aeruginosus | 2480482 |
| Chordata | Aves | Accipitriformes | Accipitridae | Circus | Circus cyaneus | 2480487 |
| Chordata | Aves | Accipitriformes | Accipitridae | Circus | Circus macrourus | 2480491 |
| Chordata | Aves | Accipitriformes | Accipitridae | Circus | Circus pygargus | 2480495 |
| Chordata | Aves | Pteroclidiformes | Pteroclididae | Syrrhaptes | Syrrhaptes paradoxus | 2480208 |
| Chordata | Aves | Otidiformes | Otididae | Chlamydotis | Chlamydotis macqueenii | 2474863 |
| Chordata | Aves | Charadriiformes | Laridae | Larus | Larus michahellis | 9413670 |
| Chordata | Aves | Procellariiformes | Procellariidae | Puffinus | Puffinus lherminieri | 5229361 |
| Chordata | Aves | Strigiformes | Strigidae | Ketupa | Ketupa zeylonensis | 5232229 |
| Chordata | Aves | Coraciiformes | Meropidae | Merops | Merops persicus | 2475419 |
| Chordata | Aves | Piciformes | Picidae | Dendrocopos | Dendrocopos leucotos | 2477927 |
| Chordata | Aves | Piciformes | Picidae | Picus | Picus sharpei | 9029556 |
| Chordata | Aves | Passeriformes | Corvidae | Cyanopica | Cyanopica cooki | 5844936 |
| Chordata | Aves | Passeriformes | Corvidae | Corvus | Corvus cornix | 2482515 |
| Chordata | Aves | Passeriformes | Paridae | Cyanistes | Cyanistes teneriffae | 7341849 |
| Chordata | Aves | Passeriformes | Alaudidae | Melanocorypha | Melanocorypha yeltoniensis | 2490614 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Iduna | Iduna rama | 5739357 |
| Chordata | Aves | Passeriformes | Acrocephalidae | Iduna | Iduna opaca | 5739355 |
| Chordata | Aves | Passeriformes | Pycnonotidae | Pycnonotus | Pycnonotus barbatus | 2486147 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus orientalis | 4408827 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus sindianus | 7341578 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus canariensis | 7341593 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus ibericus | 7341585 |
| Chordata | Aves | Passeriformes | Phylloscopidae | Phylloscopus | Phylloscopus nitidus | 9584952 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia crassirostris | 7342065 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia subalpina | 8381984 |
| Chordata | Aves | Passeriformes | Sylviidae | Sylvia | Sylvia balearica | 5845735 |
| Chordata | Aves | Passeriformes | Leiothrichidae | Turdoides | Argya altirostris | 2493316 |
| Chordata | Aves | Passeriformes | Regulidae | Regulus | Regulus madeirensis | 4408806 |
| Chordata | Aves | Passeriformes | Turdidae | Turdus | Turdus atrogularis | 4408790 |
| Chordata | Aves | Passeriformes | Muscicapidae | Ficedula | Ficedula albicilla | 5788886 |
| Chordata | Aves | Passeriformes | Muscicapidae | Saxicola | Saxicola maurus | 5846290 |
| Chordata | Aves | Passeriformes | Muscicapidae | Oenanthe | Oenanthe xanthoprymna | 5231239 |
| Chordata | Aves | Passeriformes | Motacillidae | Anthus | Anthus gustavi | 2490281 |
| Chordata | Aves | Passeriformes | Fringillidae | Carduelis | Carduelis corsicana | 4408961 |
| Chordata | Mammalia | Rodentia | Muridae | Acomys | Acomys cilicicus | 2438254 |
| Chordata | Mammalia | Rodentia | Cricetidae | Allocricetulus | Allocricetulus eversmanni | 2439190 |
| Chordata | Mammalia | Lagomorpha | Leporidae | Lepus | Lepus granatensis | 2436790 |
| Chordata | Mammalia | Lagomorpha | Leporidae | Lepus | Lepus timidus | 2436756 |
| Chordata | Mammalia | Lagomorpha | Leporidae | Lepus | Lepus tolai | 2436778 |
| Chordata | Mammalia | Carnivora | Mustelidae | Lutra | Lutra lutra | 2433753 |
| Chordata | Mammalia | Carnivora | Felidae | Lynx | Lynx lynx | 2435240 |
| Chordata | Mammalia | Carnivora | Felidae | Lynx | Lynx pardinus | 2435261 |
| Chordata | Mammalia | Rodentia | Sciuridae | Marmota | Marmota bobak | 2437375 |
| Chordata | Mammalia | Rodentia | Sciuridae | Marmota | Marmota marmota | 2437377 |
| Chordata | Mammalia | Carnivora | Canidae | Vulpes | Alopex lagopus | 5219303 |
| Chordata | Mammalia | Carnivora | Mustelidae | Martes | Martes foina | 5218887 |
| Chordata | Mammalia | Carnivora | Mustelidae | Martes | Martes martes | 5218878 |
| Chordata | Mammalia | Carnivora | Mustelidae | Martes | Martes zibellina | 5218826 |
| Chordata | Mammalia | Carnivora | Mustelidae | Meles | Meles leucurus | 2433868 |
| Chordata | Mammalia | Carnivora | Mustelidae | Meles | Meles meles | 2433875 |
| Chordata | Mammalia | Rodentia | Muridae | Meriones | Meriones crassus | 5219724 |
| Chordata | Mammalia | Rodentia | Muridae | Meriones | Meriones dahli | 5219717 |
| Chordata | Mammalia | Rodentia | Muridae | Meriones | Meriones libycus | 5219725 |
| Chordata | Mammalia | Rodentia | Muridae | Meriones | Meriones meridianus | 5219713 |
| Chordata | Mammalia | Rodentia | Muridae | Meriones | Meriones persicus | 5219719 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus agrarius | 2437761 |
| Chordata | Mammalia | Rodentia | Muridae | Meriones | Meriones tamariscinus | 5219727 |
| Chordata | Mammalia | Rodentia | Muridae | Meriones | Meriones tristrami | 5219715 |
| Chordata | Mammalia | Rodentia | Muridae | Meriones | Meriones vinogradovi | 5219716 |
| Chordata | Mammalia | Rodentia | Cricetidae | Mesocricetus | Mesocricetus auratus | 2438482 |
| Chordata | Mammalia | Rodentia | Cricetidae | Mesocricetus | Mesocricetus brandti | 2438484 |
| Chordata | Mammalia | Rodentia | Cricetidae | Mesocricetus | Mesocricetus newtoni | 2438483 |
| Chordata | Mammalia | Rodentia | Cricetidae | Mesocricetus | Mesocricetus raddei | 2438481 |
| Chordata | Mammalia | Rodentia | Muridae | Micromys | Micromys minutus | 5219833 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus agrestis | 2438616 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus anatolicus | 4265014 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus alpicola | 2437769 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus arvalis | 2438606 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus brachycercus | 4265022 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus cabrerae | 2438668 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus daghestanicus | 2438633 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus dogramacii | 4265011 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus duodecimcostatus | 2438619 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus felteni | 2438597 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus gerbei | 2438662 |
| Chordata | Mammalia | Rodentia | Cricetidae | Lasiopodomys | Lasiopodomys gregalis | 11540999 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus epimelas | 4264851 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus guentheri | 2438628 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus liechtensteini | 4265021 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus lusitanicus | 2438669 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus majori | 2438654 |
| Chordata | Mammalia | Rodentia | Cricetidae | Alexandromys | Alexandromys middendorffii | 11534627 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus multiplex | 2438647 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Alexandromys oeconomus | 2438635 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus savii | 2438650 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus flavicollis | 2437756 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus socialis | 2438670 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus subterraneus | 2438660 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus tatricus | 2438637 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus thomasi | 2438625 |
| Chordata | Mammalia | Chiroptera | Miniopteridae | Miniopterus | Miniopterus schreibersii | 9796816 |
| Chordata | Mammalia | Rodentia | Muridae | Mus | Mus cypriacus | 5786703 |
| Chordata | Mammalia | Rodentia | Muridae | Mus | Mus macedonicus | 2438783 |
| Chordata | Mammalia | Rodentia | Muridae | Mus | Mus musculus | 7429082 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus hyrcanicus | 5219733 |
| Chordata | Mammalia | Rodentia | Muridae | Mus | Mus spicilegus | 2438805 |
| Chordata | Mammalia | Rodentia | Muridae | Mus | Mus spretus | 2438793 |
| Chordata | Mammalia | Rodentia | Gliridae | Muscardinus | Muscardinus avellanarius | 2439658 |
| Chordata | Mammalia | Carnivora | Mustelidae | Mustela | Mustela erminea | 5219019 |
| Chordata | Mammalia | Carnivora | Mustelidae | Mustela | Mustela eversmanii | 5219057 |
| Chordata | Mammalia | Carnivora | Mustelidae | Mustela | Mustela lutreola | 5219007 |
| Chordata | Mammalia | Carnivora | Mustelidae | Mustela | Mustela nivalis | 5218987 |
| Chordata | Mammalia | Carnivora | Mustelidae | Mustela | Mustela putorius | 5218911 |
| Chordata | Mammalia | Carnivora | Mustelidae | Mustela | Mustela sibirica | 5218972 |
| Chordata | Mammalia | Rodentia | Cricetidae | Myodes | Myodes glareolus | 5706764 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus mystacinus | 2437773 |
| Chordata | Mammalia | Rodentia | Cricetidae | Myodes | Craseomys rufocanus | 5706769 |
| Chordata | Mammalia | Rodentia | Cricetidae | Myodes | Myodes rutilus | 5706762 |
| Chordata | Mammalia | Rodentia | Gliridae | Myomimus | Myomimus roachi | 2439663 |
| Chordata | Mammalia | Rodentia | Gliridae | Myomimus | Myomimus setzeri | 2439662 |
| Chordata | Mammalia | Rodentia | Cricetidae | Myopus | Myopus schisticolor | 2439358 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis alcathoe | 4266346 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis bechsteinii | 2432427 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis blythii | 2432414 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis brandtii | 7261875 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus ponticus | 5219740 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis capaccinii | 2432430 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis dasycneme | 2432452 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis daubentonii | 2432439 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis emarginatus | 2432470 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis myotis | 2432416 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis mystacinus | 9754263 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis nattereri | 2432389 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis punicus | 4266337 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus sylvaticus | 2437760 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis schaubi | 2432417 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Neomys | Neomys anomalus | 7825895 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Neomys | Neomys fodiens | 2435767 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Neomys | Neomys teres | 5219474 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Nyctalus | Nyctalus azoreum | 5218523 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Nyctalus | Nyctalus lasiopterus | 5218525 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Nyctalus | Nyctalus leisleri | 5218522 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Nyctalus | Nyctalus noctula | 5218524 |
| Chordata | Mammalia | Lagomorpha | Ochotonidae | Ochotona | Ochotona hyperborea | 2437002 |
| Chordata | Mammalia | Rodentia | Muridae | Acomys | Acomys minous | 2438264 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus uralensis | 2437775 |
| Chordata | Mammalia | Lagomorpha | Ochotonidae | Ochotona | Ochotona pusilla | 2437017 |
| Chordata | Mammalia | Lagomorpha | Leporidae | Oryctolagus | Oryctolagus cuniculus | 2436940 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Ovis | Ovis gmelini | 9542814 |
| Chordata | Mammalia | Carnivora | Felidae | Panthera | Panthera pardus | 5219436 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Pipistrellus | Pipistrellus kuhlii | 5218464 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Pipistrellus | Pipistrellus maderensis | 5218476 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Pipistrellus | Pipistrellus nathusii | 5218471 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Pipistrellus | Pipistrellus pipistrellus | 5218465 |
| Chordata | Mammalia | Rodentia | Muridae | Apodemus | Apodemus witherbyi | 5706747 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Pipistrellus | Pipistrellus pygmaeus | 5707150 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Plecotus | Plecotus auritus | 5218507 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Plecotus | Plecotus austriacus | 5739437 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Plecotus | Plecotus kolombatovici | 5739445 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Plecotus | Plecotus macrobullaris | 5787719 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Plecotus | Plecotus sardus | 5739436 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Plecotus | Plecotus teneriffae | 5218519 |
| Chordata | Mammalia | Rodentia | Cricetidae | Prometheomys | Prometheomys schaposchnikowi | 2438887 |
| Chordata | Mammalia | Rodentia | Sciuridae | Pteromys | Pteromys volans | 2437258 |
| Chordata | Mammalia | Rodentia | Cricetidae | Arvicola | Arvicola amphibius | 4265185 |
| Chordata | Mammalia | Rodentia | Dipodidae | Pygeretmus | Pygeretmus pumilio | 2439433 |
| Chordata | Mammalia | Artiodactyla | Cervidae | Rangifer | Rangifer tarandus | 5220114 |
| Chordata | Mammalia | Chiroptera | Rhinolophidae | Rhinolophus | Rhinolophus blasii | 2432666 |
| Chordata | Mammalia | Chiroptera | Rhinolophidae | Rhinolophus | Rhinolophus euryale | 2432621 |
| Chordata | Mammalia | Chiroptera | Rhinolophidae | Rhinolophus | Rhinolophus ferrumequinum | 2432655 |
| Chordata | Mammalia | Chiroptera | Rhinolophidae | Rhinolophus | Rhinolophus hipposideros | 2432614 |
| Chordata | Mammalia | Chiroptera | Rhinolophidae | Rhinolophus | Rhinolophus mehelyi | 2432667 |
| Chordata | Mammalia | Rodentia | Cricetidae | Arvicola | Arvicola sapidus | 2438492 |
| Chordata | Mammalia | Chiroptera | Pteropodidae | Rousettus | Rousettus aegyptiacus | 2432953 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Rupicapra | Rupicapra pyrenaica | 5220171 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Rupicapra | Rupicapra rupicapra | 5220170 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Saiga | Saiga tatarica | 2441067 |
| Chordata | Mammalia | Rodentia | Sciuridae | Sciurus | Sciurus anomalus | 8243249 |
| Chordata | Mammalia | Rodentia | Sciuridae | Sciurus | Sciurus vulgaris | 8211070 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista armenica | 2439444 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista betulina | 2439449 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista caucasica | 2439440 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista kazbegica | 2439450 |
| Chordata | Mammalia | Rodentia | Cricetidae | Arvicola | Arvicola monticola | 4265186 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista kluchorica | 2439441 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista strandi | 2439448 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista subtilis | 2439439 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex alpinus | 2435986 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex antinorii | 2435998 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex araneus | 8316400 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex caecutiens | 2435994 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex coronatus | 2436090 |
| Chordata | Mammalia | Erinaceomorpha | Erinaceidae | Atelerix | Atelerix algirus | 2437136 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex granarius | 2436071 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex isodon | 2436016 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex minutissimus | 2435959 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex minutus | 7571319 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex raddei | 2436036 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex samniticus | 2436103 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex satunini | 2436072 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex tundrensis | 2435975 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex volnuchini | 2436092 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Spalax arenarius | 2438884 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Barbastella | Barbastella barbastellus | 2432582 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Nannospalax ehrenbergi | 4264694 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Spalax giganteus | 2438882 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Spalax graecus | 2438885 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Nannospalax leucodon | 4264696 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Spalax zemni | 2438881 |
| Chordata | Mammalia | Rodentia | Sciuridae | Spermophilus | Spermophilus citellus | 2437304 |
| Chordata | Mammalia | Rodentia | Sciuridae | Spermophilus | Spermophilus fulvus | 2437306 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Barbastella | Barbastella caspica | 5787711 |
| Chordata | Mammalia | Rodentia | Sciuridae | Spermophilus | Spermophilus major | 2437320 |
| Chordata | Mammalia | Rodentia | Sciuridae | Spermophilus | Spermophilus musicus | 2437308 |
| Chordata | Mammalia | Rodentia | Sciuridae | Spermophilus | Spermophilus pygmaeus | 2437322 |
| Chordata | Mammalia | Rodentia | Sciuridae | Spermophilus | Spermophilus suslicus | 2437323 |
| Chordata | Mammalia | Rodentia | Sciuridae | Spermophilus | Spermophilus taurensis | 9040543 |
| Chordata | Mammalia | Rodentia | Sciuridae | Spermophilus | Spermophilus xanthoprymnus | 2437301 |
| Chordata | Mammalia | Rodentia | Dipodidae | Stylodipus | Stylodipus telum | 2439453 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Suncus | Suncus etruscus | 2435506 |
| Chordata | Mammalia | Artiodactyla | Suidae | Sus | Sus scrofa | 7705930 |
| Chordata | Mammalia | Chiroptera | Molossidae | Tadarida | Tadarida teniotis | 2433009 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Bison | Bison bonasus | 2441184 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa caeca | 8347063 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa caucasica | 2436136 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa davidiana | 2436158 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa europaea | 7872906 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa levantis | 2436131 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa occidentalis | 2436152 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa romana | 2436140 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa stankovici | 2436128 |
| Chordata | Mammalia | Rodentia | Sciuridae | Tamias | Tamias sibiricus | 2437450 |
| Chordata | Mammalia | Rodentia | Muridae | Tatera | Tatera indica | 2438134 |
| Chordata | Mammalia | Rodentia | Calomyscidae | Calomyscus | Calomyscus urartensis | 2439354 |
| Chordata | Mammalia | Carnivora | Ursidae | Ursus | Ursus arctos | 2433433 |
| Chordata | Mammalia | Carnivora | Ursidae | Ursus | Ursus maritimus | 2433451 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Vespertilio | Vespertilio murinus | 2432564 |
| Chordata | Mammalia | Carnivora | Mustelidae | Vormela | Vormela peregusna | 2433709 |
| Chordata | Mammalia | Carnivora | Canidae | Vulpes | Vulpes corsac | 5219293 |
| Chordata | Mammalia | Carnivora | Canidae | Vulpes | Vulpes vulpes | 5219243 |
| Chordata | Mammalia | Rodentia | Muridae | Acomys | Acomys nesiotes | 2438261 |
| Chordata | Mammalia | Carnivora | Canidae | Canis | Canis aureus | 5219219 |
| Chordata | Mammalia | Carnivora | Canidae | Canis | Canis lupus | 5219173 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Capra | Capra aegagrus | 4262706 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Capra | Capra caucasica | 2441051 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Capra | Capra cylindricornis | 4262708 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Capra | Capra ibex | 2441055 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Capra | Capra pyrenaica | 2441054 |
| Chordata | Mammalia | Artiodactyla | Cervidae | Capreolus | Capreolus capreolus | 5220126 |
| Chordata | Mammalia | Artiodactyla | Cervidae | Capreolus | Capreolus pygargus | 5220127 |
| Chordata | Mammalia | Artiodactyla | Cervidae | Alces | Alces alces | 2440940 |
| Chordata | Mammalia | Rodentia | Castoridae | Castor | Castor fiber | 4409131 |
| Chordata | Mammalia | Artiodactyla | Cervidae | Cervus | Cervus elaphus | 2440958 |
| Chordata | Mammalia | Rodentia | Cricetidae | Chionomys | Chionomys gud | 2438160 |
| Chordata | Mammalia | Rodentia | Cricetidae | Chionomys | Chionomys nivalis | 2438162 |
| Chordata | Mammalia | Rodentia | Cricetidae | Chionomys | Chionomys roberti | 2438161 |
| Chordata | Mammalia | Rodentia | Cricetidae | Cricetulus | Cricetulus migratorius | 2438772 |
| Chordata | Mammalia | Rodentia | Cricetidae | Cricetus | Cricetus cricetus | 2438704 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura arispa | 2435612 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura canariensis | 2435551 |
| Chordata | Mammalia | Rodentia | Dipodidae | Allactaga | Scarturus elater | 2439489 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura caspica | 2435744 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura leucodon | 2435723 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura pachyura | 5787823 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura russula | 2435654 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura sicula | 2435553 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura suaveolens | 2435687 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura zimmermanni | 2435665 |
| Chordata | Mammalia | Artiodactyla | Cervidae | Dama | Dama dama | 5220136 |
| Chordata | Mammalia | Rodentia | Dipodidae | Scarturus | Scarturus euphraticus | 11507230 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Desmana | Desmana moschata | 2436117 |
| Chordata | Mammalia | Rodentia | Cricetidae | Dicrostonyx | Dicrostonyx torquatus | 2438384 |
| Chordata | Mammalia | Rodentia | Cricetidae | Dinaromys | Dinaromys bogdanovi | 2438502 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Diplomesodon | Diplomesodon pulchellus | 2435917 |
| Chordata | Mammalia | Rodentia | Dipodidae | Dipus | Dipus sagitta | 2439463 |
| Chordata | Mammalia | Rodentia | Gliridae | Dryomys | Dryomys laniger | 2439686 |
| Chordata | Mammalia | Rodentia | Gliridae | Dryomys | Dryomys nitedula | 2439688 |
| Chordata | Mammalia | Rodentia | Gliridae | Eliomys | Eliomys melanurus | 2439684 |
| Chordata | Mammalia | Rodentia | Gliridae | Eliomys | Eliomys quercinus | 2439683 |
| Chordata | Mammalia | Rodentia | Cricetidae | Ellobius | Ellobius lutescens | 2438762 |
| Chordata | Mammalia | Rodentia | Dipodidae | Allactaga | Allactaga major | 2439483 |
| Chordata | Mammalia | Rodentia | Cricetidae | Ellobius | Ellobius talpinus | 2438760 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Eptesicus | Eptesicus nilssonii | 7261816 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Eptesicus | Eptesicus serotinus | 2432359 |
| Chordata | Mammalia | Erinaceomorpha | Erinaceidae | Erinaceus | Erinaceus concolor | 5219624 |
| Chordata | Mammalia | Erinaceomorpha | Erinaceidae | Erinaceus | Erinaceus europaeus | 5219616 |
| Chordata | Mammalia | Erinaceomorpha | Erinaceidae | Erinaceus | Erinaceus roumanicus | 5219618 |
| Chordata | Mammalia | Carnivora | Felidae | Felis | Felis chaus | 2435066 |
| Chordata | Mammalia | Carnivora | Felidae | Felis | Felis silvestris | 7964291 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Galemys | Galemys pyrenaicus | 5219516 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Gazella | Gazella subgutturosa | 5220146 |
| Chordata | Mammalia | Carnivora | Viverridae | Genetta | Genetta genetta | 5219362 |
| Chordata | Mammalia | Rodentia | Muridae | Dipodillus | Gerbillus dasyurus | 4264993 |
| Chordata | Mammalia | Rodentia | Gliridae | Glis | Glis glis | 5706486 |
| Chordata | Mammalia | Carnivora | Mustelidae | Gulo | Gulo gulo | 5219073 |
| Chordata | Mammalia | Erinaceomorpha | Erinaceidae | Hemiechinus | Hemiechinus auritus | 2437125 |
| Chordata | Mammalia | Carnivora | Herpestidae | Herpestes | Herpestes ichneumon | 2434255 |
| Chordata | Mammalia | Carnivora | Hyaenidae | Hyaena | Hyaena hyaena | 5218777 |
| Chordata | Mammalia | Rodentia | Dipodidae | Scarturus | Scarturus williamsi | 10954088 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Hypsugo | Hypsugo savii | 7261861 |
| Chordata | Mammalia | Rodentia | Hystricidae | Hystrix | Hystrix cristata | 5219888 |
| Chordata | Mammalia | Rodentia | Hystricidae | Hystrix | Hystrix indica | 5219884 |
| Chordata | Mammalia | Rodentia | Cricetidae | Lagurus | Lagurus lagurus | 2439131 |
| Chordata | Mammalia | Rodentia | Cricetidae | Lemmus | Lemmus lemmus | 5219840 |
| Chordata | Mammalia | Rodentia | Cricetidae | Lemmus | Lemmus sibiricus | 5219835 |
| Chordata | Mammalia | Lagomorpha | Leporidae | Lepus | Lepus capensis | 2436736 |
| Chordata | Mammalia | Lagomorpha | Leporidae | Lepus | Lepus castroviejoi | 2436788 |
| Chordata | Mammalia | Lagomorpha | Leporidae | Lepus | Lepus corsicanus | 2436789 |
| Chordata | Mammalia | Lagomorpha | Leporidae | Lepus | Lepus europaeus | 7952072 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Sorex | Sorex daphaenodon | 2436000 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Neomys | Neomys milleri | 8160177 |
| Chordata | Mammalia | Soricomorpha | Soricidae | Crocidura | Crocidura gueldenstaedtii | 2435687 |
| Chordata | Mammalia | Soricomorpha | Talpidae | Talpa | Talpa aquitania | 9835106 |
| Chordata | Mammalia | Chiroptera | Emballonuridae | Taphozous | Taphozous nudiventris | 5218737 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Eptesicus | Eptesicus ognevi | 9065533 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Eptesicus | Eptesicus anatolicus | 5787592 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Eptesicus | Eptesicus isabellinus | 5787585 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Plecotus | Plecotus gaisleri | 10893276 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Otonycteris | Otonycteris hemprichii | 2432382 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis davidii | 4266349 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis crypticus | 9918569 |
| Chordata | Mammalia | Chiroptera | Vespertilionidae | Myotis | Myotis escalerai | 8181305 |
| Chordata | Mammalia | Chiroptera | Miniopteridae | Miniopterus | Miniopterus pallidus | 7500714 |
| Chordata | Mammalia | Carnivora | Felidae | Caracal | Caracal caracal | 2435010 |
| Chordata | Mammalia | Carnivora | Felidae | Felis | Otocolobus manul | 2435023 |
| Chordata | Mammalia | Artiodactyla | Bovidae | Gazella | Gazella gazella | 5220154 |
| Chordata | Mammalia | Rodentia | Gliridae | Dryomys | Dryomys aspromontis | 2439685 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista trizona | 8328725 |
| Chordata | Mammalia | Rodentia | Dipodidae | Sicista | Sicista loriger | 2439439 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Spalax antiquus | 10753894 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Spalax microphtalmus | 2438883 |
| Chordata | Mammalia | Rodentia | Spalacidae | Spalax | Nannospalax xanthodon | 10913440 |
| Chordata | Mammalia | Rodentia | Cricetidae | Arvicola | Arvicola italicus | 11801822 |
| Chordata | Mammalia | Rodentia | Cricetidae | Chionomys | Chionomys lasistanius | 2438160 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus lavernedii | 10517059 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus rozianus | 2438616 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus hartingi | 2438628 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus obscurus | 2438606 |
| Chordata | Mammalia | Rodentia | Cricetidae | Microtus | Microtus mystacinus | 2438606 |
| Chordata | Mammalia | Rodentia | Muridae | Nesokia | Nesokia indica | 2438985 |
| Chordata | Squamata | Phyllodactylidae | Asaccus | Asaccus barani | 6066757 | |
| Chordata | Squamata | Gekkonidae | Mediodactylus | Mediodactylus danilewskii | 11165726 | |
| Chordata | Squamata | Gekkonidae | Mediodactylus | Mediodactylus oertzeni | 11199676 | |
| Chordata | Squamata | Gekkonidae | Mediodactylus | Mediodactylus orientalis | 11153404 | |
| Chordata | Squamata | Lacertidae | Anatololacerta | Anatololacerta pelasgiana | 9565497 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia bithynica | 9189529 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta diplochondrodes | 11189228 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis cretensis | 5789532 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis guadarramae | 9258179 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis ionicus | 11180835 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis levendis | 5789530 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis virescens | 9323087 | |
| Chordata | Squamata | Lacertidae | Psammodromus | Psammodromus algirus | 2469763 | |
| Chordata | Squamata | Lacertidae | Psammodromus | Psammodromus occidentalis | 7416224 | |
| Chordata | Squamata | Lacertidae | Timon | Timon nevadensis | 9574630 | |
| Chordata | Squamata | Lacertidae | Timon | Timon tangitanus | 2468540 | |
| Chordata | Squamata | Scincidae | Chalcides | Chalcides coeruleopunctatus | 8247946 | |
| Chordata | Squamata | Anguidae | Anguis | Anguis colchica | 8356845 | |
| Chordata | Squamata | Anguidae | Anguis | Anguis graeca | 8279309 | |
| Chordata | Squamata | Anguidae | Anguis | Anguis veronensis | 7567645 | |
| Chordata | Squamata | Blanidae | Blanus | Blanus aporus | 4821422 | |
| Chordata | Squamata | Blanidae | Blanus | Blanus vandellii | 9305963 | |
| Chordata | Squamata | Boidae | Eryx | Eryx tataricus | 2465319 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis collaris | 2453265 | |
| Chordata | Squamata | Colubridae | Elaphe | Elaphe urartica | 10287547 | |
| Chordata | Squamata | Colubridae | Natrix | Natrix astreptophora | 9313254 | |
| Chordata | Squamata | Colubridae | Natrix | Natrix helvetica | 9350124 | |
| Chordata | Squamata | Psammophiidae | Malpolon | Malpolon insignitus | 5959662 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera anatolica | 9598629 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera barani | 5960851 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera graeca | 9012191 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera magnifica | 5960856 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera nikolskii | 5960875 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera orlovi | 5960885 | |
| Chordata | Squamata | Scincidae | Ablepharus | Ablepharus bivittatus | 2461305 | |
| Chordata | Squamata | Lacertidae | Algyroides | Algyroides moreoticus | 2469367 | |
| Chordata | Squamata | Lacertidae | Hellenolacerta | Hellenolacerta graeca | 5789567 | |
| Chordata | Squamata | Gekkonidae | Hemidactylus | Hemidactylus turcicus | 5221528 | |
| Chordata | Squamata | Colubridae | Hemorrhois | Hemorrhois hippocrepis | 2459023 | |
| Chordata | Squamata | Colubridae | Hierophis | Dolichophis cypriensis | 2457223 | |
| Chordata | Squamata | Colubridae | Hierophis | Hierophis gemonensis | 2458928 | |
| Chordata | Squamata | Colubridae | Hierophis | Hierophis viridiflavus | 2458916 | |
| Chordata | Squamata | Lacertidae | Iberolacerta | Iberolacerta aranica | 7837038 | |
| Chordata | Squamata | Lacertidae | Iberolacerta | Iberolacerta aurelioi | 2468520 | |
| Chordata | Squamata | Lacertidae | Iberolacerta | Iberolacerta bonnali | 2468525 | |
| Chordata | Squamata | Lacertidae | Algyroides | Algyroides nigropunctatus | 2469363 | |
| Chordata | Squamata | Lacertidae | Iberolacerta | Iberolacerta cyreni | 2468503 | |
| Chordata | Squamata | Lacertidae | Iberolacerta | Iberolacerta galani | 2468522 | |
| Chordata | Squamata | Lacertidae | Iberolacerta | Iberolacerta horvathi | 2468506 | |
| Chordata | Squamata | Lacertidae | Iberolacerta | Iberolacerta martinezricai | 2468518 | |
| Chordata | Squamata | Lacertidae | Iberolacerta | Iberolacerta monticola | 2468511 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta agilis | 6159223 | |
| Chordata | Squamata | Lacertidae | Anatololacerta | Anatololacerta anatolica | 5789536 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta bilineata | 6159243 | |
| Chordata | Squamata | Lacertidae | Apathya | Apathya cappadocica | 2469406 | |
| Chordata | Squamata | Lacertidae | Phoenicolacerta | Phoenicolacerta cyanisparsa | 5789551 | |
| Chordata | Squamata | Gekkonidae | Alsophylax | Alsophylax pipiens | 2444983 | |
| Chordata | Squamata | Lacertidae | Anatololacerta | Anatololacerta danfordi | 5789533 | |
| Chordata | Squamata | Lacertidae | Teira | Teira dugesii | 2468700 | |
| Chordata | Squamata | Lacertidae | Phoenicolacerta | Phoenicolacerta laevis | 5789554 | |
| Chordata | Squamata | Lacertidae | Timon | Timon lepidus | 2468542 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta media | 6159264 | |
| Chordata | Squamata | Lacertidae | Dinarolacerta | Dinarolacerta mosorensis | 5789566 | |
| Chordata | Squamata | Lacertidae | Dinarolacerta | Dinarolacerta montenegrina | 8094812 | |
| Chordata | Squamata | Lacertidae | Dalmatolacerta | Dalmatolacerta oxycephala | 5789555 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta pamphylica | 6159256 | |
| Chordata | Squamata | Anguidae | Anguis | Anguis cephallonica | 2468088 | |
| Chordata | Squamata | Lacertidae | Parvilacerta | Parvilacerta parva | 5226786 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta schreiberi | 6159196 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta strigata | 6159238 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta trilineata | 6159197 | |
| Chordata | Squamata | Lacertidae | Phoenicolacerta | Phoenicolacerta troodica | 5789552 | |
| Chordata | Squamata | Lacertidae | Lacerta | Lacerta viridis | 6159273 | |
| Chordata | Squamata | Lacertidae | Zootoca | Zootoca vivipara | 9702100 | |
| Chordata | Squamata | Agamidae | Paralaudakia | Paralaudakia caucasia | 7376912 | |
| Chordata | Squamata | Agamidae | Stellagama | Stellagama stellio | 7504253 | |
| Chordata | Squamata | Anguidae | Anguis | Anguis fragilis | 2468095 | |
| Chordata | Squamata | Leptotyphlopidae | Myriopholis | Myriopholis macrorhyncha | 8050746 | |
| Chordata | Squamata | Scincidae | Heremites | Heremites auratus | 11068456 | |
| Chordata | Squamata | Scincidae | Heremites | Heremites vittatus | 10910391 | |
| Chordata | Squamata | Colubridae | Macroprotodon | Macroprotodon brevis | 2455958 | |
| Chordata | Squamata | Colubridae | Macroprotodon | Macroprotodon cucullatus | 7881183 | |
| Chordata | Squamata | Viperidae | Macrovipera | Macrovipera lebetina | 2444716 | |
| Chordata | Squamata | Viperidae | Macrovipera | Macrovipera schweizeri | 2444698 | |
| Chordata | Squamata | Psammophiidae | Malpolon | Malpolon monspessulanus | 8796529 | |
| Chordata | Testudines | Geoemydidae | Mauremys | Mauremys caspica | 2443374 | |
| Chordata | Testudines | Geoemydidae | Mauremys | Mauremys leprosa | 2443341 | |
| Chordata | Squamata | Lacertidae | Archaeolacerta | Archaeolacerta bedriagae | 9107061 | |
| Chordata | Testudines | Geoemydidae | Mauremys | Mauremys rivulata | 2443384 | |
| Chordata | Squamata | Viperidae | Montivipera | Montivipera raddei | 5789505 | |
| Chordata | Squamata | Viperidae | Montivipera | Montivipera wagneri | 5789501 | |
| Chordata | Squamata | Viperidae | Montivipera | Montivipera xanthina | 4820419 | |
| Chordata | Squamata | Colubridae | Natrix | Natrix maura | 6161340 | |
| Chordata | Squamata | Colubridae | Natrix | Natrix natrix | 6161347 | |
| Chordata | Squamata | Colubridae | Natrix | Natrix tessellata | 6161327 | |
| Chordata | Squamata | Scincidae | Ophiomorus | Ophiomorus punctatissimus | 2462191 | |
| Chordata | Squamata | Lacertidae | Ophisops | Ophisops elegans | 2469702 | |
| Chordata | Squamata | Agamidae | Phrynocephalus | Phrynocephalus guttatus | 8959353 | |
| Chordata | Squamata | Phyllodactylidae | Asaccus | Asaccus elisae | 2447277 | |
| Chordata | Squamata | Agamidae | Phrynocephalus | Phrynocephalus helioscopus | 2466398 | |
| Chordata | Squamata | Agamidae | Phrynocephalus | Phrynocephalus mystaceus | 9152383 | |
| Chordata | Squamata | Agamidae | Phrynocephalus | Phrynocephalus persicus | 2466313 | |
| Chordata | Squamata | Colubridae | Platyceps | Platyceps najadum | 5223135 | |
| Chordata | Squamata | Lacertidae | Iranolacerta | Iranolacerta brandtii | 5789521 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis liolepis | 7444957 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis bocagei | 2469143 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis carbonelli | 2469141 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis erhardii | 2469075 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis filfolensis | 2469148 | |
| Chordata | Squamata | Blanidae | Blanus | Blanus cinereus | 2473222 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis gaigeae | 2468994 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis hispanicus | 9019391 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis lilfordi | 2469155 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis melisellensis | 2468999 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis milensis | 2469118 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis muralis | 2469188 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis peloponnesiacus | 2469022 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis pityusensis | 2469028 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis raffonei | 2469122 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis siculus | 9185677 | |
| Chordata | Squamata | Blanidae | Blanus | Blanus strauchi | 2473227 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis tauricus | 2469065 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis tiliguerta | 2469126 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis vaucheri | 2469139 | |
| Chordata | Squamata | Lacertidae | Podarcis | Podarcis waglerianus | 8145436 | |
| Chordata | Squamata | Lacertidae | Psammodromus | Psammodromus hispanicus | 7917188 | |
| Chordata | Squamata | Anguidae | Pseudopus | Pseudopus apodus | 2468350 | |
| Chordata | Testudines | Trionychidae | Rafetus | Rafetus euphraticus | 2442640 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia chlorogaster | 2469411 | |
| Chordata | Squamata | Phyllodactylidae | Tarentola | Tarentola angustimentalis | 2445033 | |
| Chordata | Squamata | Scincidae | Chalcides | Chalcides bedriagai | 8974077 | |
| Chordata | Squamata | Phyllodactylidae | Tarentola | Tarentola bischoffi | 2445052 | |
| Chordata | Squamata | Phyllodactylidae | Tarentola | Tarentola boettgeri | 2445052 | |
| Chordata | Squamata | Phyllodactylidae | Tarentola | Tarentola delalandii | 8994891 | |
| Chordata | Squamata | Phyllodactylidae | Tarentola | Tarentola gomerensis | 2445039 | |
| Chordata | Squamata | Phyllodactylidae | Tarentola | Tarentola mauritanica | 2445034 | |
| Chordata | Squamata | Colubridae | Telescopus | Telescopus fallax | 2457466 | |
| Chordata | Testudines | Testudinidae | Testudo | Testudo graeca | 2441461 | |
| Chordata | Testudines | Testudinidae | Testudo | Testudo hermanni | 2441454 | |
| Chordata | Testudines | Emydidae | Emys | Emys trinacris | 5220531 | |
| Chordata | Testudines | Testudinidae | Testudo | Testudo marginata | 2441502 | |
| Chordata | Squamata | Scincidae | Ablepharus | Ablepharus budaki | 8011236 | |
| Chordata | Squamata | Agamidae | Trapelus | Trapelus agilis | 9204508 | |
| Chordata | Squamata | Agamidae | Trapelus | Trapelus ruderatus | 9780750 | |
| Chordata | Testudines | Trionychidae | Trionyx | Trionyx triunguis | 5960976 | |
| Chordata | Squamata | Typhlopidae | Xerotyphlops | Xerotyphlops vermicularis | 9624852 | |
| Chordata | Squamata | Varanidae | Varanus | Varanus griseus | 9628941 | |
| Chordata | Squamata | Viperidae | Montivipera | Montivipera albizona | 5789499 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera ammodytes | 2444308 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera aspis | 2444339 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera berus | 5960862 | |
| Chordata | Squamata | Viperidae | Montivipera | Montivipera bulgardaghica | 5789507 | |
| Chordata | Squamata | Scincidae | Chalcides | Chalcides chalcides | 5225173 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera darevskii | 5960852 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera dinniki | 5960888 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera kaznakovi | 5960856 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera latastei | 5960896 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera pontica | 5960903 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera transcaucasiana | 5960866 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera seoanei | 5960884 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera ursinii | 9807306 | |
| Chordata | Squamata | Colubridae | Dolichophis | Dolichophis caspius | 2457224 | |
| Chordata | Squamata | Scincidae | Chalcides | Chalcides ocellatus | 5225137 | |
| Chordata | Squamata | Chamaeleonidae | Chamaeleo | Chamaeleo africanus | 2448980 | |
| Chordata | Squamata | Colubridae | Dolichophis | Dolichophis jugularis | 2457232 | |
| Chordata | Squamata | Colubridae | Hemorrhois | Hemorrhois nummifer | 2459015 | |
| Chordata | Squamata | Colubridae | Dolichophis | Dolichophis schmidti | 2457240 | |
| Chordata | Squamata | Colubridae | Elaphe | Elaphe sauromates | 2458386 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera renardi | 8568628 | |
| Chordata | Squamata | Gekkonidae | Stenodactylus | Stenodactylus grandiceps | 5221507 | |
| Chordata | Squamata | Lacertidae | Acanthodactylus | Acanthodactylus boskianus | 5226727 | |
| Chordata | Squamata | Scincidae | Chalcides | Chalcides sexlineatus | 5225153 | |
| Chordata | Squamata | Colubridae | Hemorrhois | Hemorrhois ravergieri | 2458997 | |
| Chordata | Squamata | Colubridae | Platyceps | Platyceps ventromaculatus | 5223128 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis persicus | 8124270 | |
| Chordata | Squamata | Colubridae | Rhynchocalamus | Rhynchocalamus melanocephalus | 2456620 | |
| Chordata | Squamata | Colubridae | Spalerosophis | Spalerosophis diadema | 9150197 | |
| Chordata | Squamata | Eublepharidae | Eublepharis | Eublepharis angramainyu | 5221290 | |
| Chordata | Squamata | Scincidae | Heremites | Heremites septemtaeniata | 10979564 | |
| Chordata | Squamata | Lacertidae | Acanthodactylus | Acanthodactylus harranensis | 5226705 | |
| Chordata | Squamata | Lacertidae | Acanthodactylus | Acanthodactylus grandis | 5226764 | |
| Chordata | Squamata | Scincidae | Chalcides | Chalcides simonyi | 5225149 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera eriwanensis | 9620019 | |
| Chordata | Squamata | Colubridae | Zamenis | Zamenis persicus | 2458397 | |
| Chordata | Squamata | Viperidae | Vipera | Vipera lotievi | 5960861 | |
| Chordata | Squamata | Lacertidae | Mesalina | Mesalina brevirostris | 2469301 | |
| Chordata | Squamata | Colubridae | Telescopus | Telescopus nigriceps | 2457476 | |
| Chordata | Squamata | Typhlopidae | Letheobia | Letheobia episcopus | 5789802 | |
| Chordata | Squamata | Colubridae | Muhtarophis | Muhtarophis barani | 9618860 | |
| Chordata | Squamata | Scincidae | Chalcides | Chalcides striatus | 5225191 | |
| Chordata | Squamata | Colubridae | Macroprotodon | Macroprotodon mauritanicus | 2455971 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis coronelloides | 5959683 | |
| Chordata | Squamata | Scincidae | Chalcides | Chalcides viridanus | 5225148 | |
| Chordata | Squamata | Chamaeleonidae | Chamaeleo | Chamaeleo chamaeleon | 2449051 | |
| Coronella austriaca | 0 | |||||
| Chordata | Squamata | Colubridae | Coronella | Coronella girondica | 5959645 | |
| Chordata | Squamata | Scincidae | Ablepharus | Ablepharus chernovi | 2461286 | |
| Chordata | Squamata | Gekkonidae | Tenuidactylus | Tenuidactylus caspius | 2447953 | |
| Chordata | Squamata | Gekkonidae | Mediodactylus | Mediodactylus heterocercus | 9510913 | |
| Chordata | Squamata | Gekkonidae | Mediodactylus | Mediodactylus kotschyi | 2447858 | |
| Chordata | Squamata | Gekkonidae | Mediodactylus | Mediodactylus russowii | 2447758 | |
| Chordata | Squamata | Gekkonidae | Cyrtopodion | Cyrtopodion scabrum | 2448051 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia alpina | 2468793 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia armeniaca | 2468847 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia bendimahiensis | 2468789 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia brauneri | 9083992 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia caucasica | 2468840 | |
| Chordata | Squamata | Scincidae | Ablepharus | Ablepharus kitaibelii | 8120403 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia clarkorum | 2468739 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia daghestanica | 2468831 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia dahli | 2468869 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia derjugini | 2468780 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia dryada | 2468838 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia lindholmi | 2468845 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia mixta | 2468735 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia parvula | 2468743 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia portschinskii | 2468751 | |
| Chordata | Squamata | Scincidae | Ablepharus | Ablepharus pannonicus | 9623013 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia praticola | 2468799 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia raddei | 8858084 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia rostombekowi | 2468770 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia rudis | 9131359 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia sapphirina | 2468863 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia saxicola | 2468822 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia unisexualis | 2468827 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia uzzelli | 2468866 | |
| Chordata | Squamata | Lacertidae | Darevskia | Darevskia valentini | 2468771 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis aurolineatus | 5959697 | |
| Chordata | Squamata | Lacertidae | Acanthodactylus | Acanthodactylus erythrurus | 5226733 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis barani | 2455740 | |
| Chordata | Squamata | Colubridae | Platyceps | Platyceps collaris | 5223125 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis coronella | 5959699 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis decemlineatus | 5959677 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis eiselti | 5959691 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis hakkariensis | 5959706 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis levantinus | 5959681 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis lineomaculatus | 5959692 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis modestus | 2455751 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis punctatolineatus | 8604614 | |
| Chordata | Squamata | Lacertidae | Acanthodactylus | Acanthodactylus schreiberi | 5226767 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis rothii | 5959675 | |
| Chordata | Squamata | Colubridae | Eirenis | Eirenis thospitis | 5959696 | |
| Chordata | Squamata | Colubridae | Elaphe | Elaphe dione | 2458400 | |
| Chordata | Squamata | Colubridae | Zamenis | Zamenis hohenackeri | 2458535 | |
| Chordata | Squamata | Colubridae | Zamenis | Zamenis lineatus | 2458382 | |
| Chordata | Squamata | Colubridae | Zamenis | Zamenis longissimus | 2458538 | |
| Chordata | Squamata | Colubridae | Elaphe | Elaphe quatuorlineata | 9099408 | |
| Chordata | Squamata | Colubridae | Zamenis | Zamenis scalaris | 9416124 | |
| Chordata | Squamata | Colubridae | Zamenis | Zamenis situla | 2458374 | |
| Chordata | Squamata | Lacertidae | Algyroides | Algyroides fitzingeri | 2469361 | |
| Chordata | Testudines | Emydidae | Emys | Emys orbicularis | 8909809 | |
| Chordata | Squamata | Lacertidae | Eremias | Eremias arguta | 2468945 | |
| Chordata | Squamata | Lacertidae | Eremias | Eremias pleskei | 2468896 | |
| Chordata | Squamata | Lacertidae | Eremias | Eremias strauchi | 2468908 | |
| Chordata | Squamata | Lacertidae | Eremias | Eremias suphani | 2468874 | |
| Chordata | Squamata | Lacertidae | Eremias | Eremias velox | 2468889 | |
| Chordata | Squamata | Boidae | Eryx | Eryx jaculus | 9595248 | |
| Chordata | Squamata | Boidae | Eryx | Eryx miliaris | 2465319 | |
| Chordata | Squamata | Lacertidae | Algyroides | Algyroides marchi | 2469356 | |
| Chordata | Squamata | Sphaerodactylidae | Euleptes | Euleptes europaea | 2446608 | |
| Chordata | Squamata | Scincidae | Eumeces | Eumeces schneiderii | 9032472 | |
| Chordata | Squamata | Lacertidae | Gallotia | Gallotia atlantica | 2468656 | |
| Chordata | Squamata | Lacertidae | Gallotia | Gallotia bravoana | 7872185 | |
| Chordata | Squamata | Lacertidae | Gallotia | Gallotia caesaris | 2468663 | |
| Chordata | Squamata | Lacertidae | Gallotia | Gallotia galloti | 8682680 | |
| Chordata | Squamata | Lacertidae | Gallotia | Gallotia intermedia | 2468688 | |
| Chordata | Squamata | Lacertidae | Gallotia | Gallotia simonyi | 2468673 | |
| Chordata | Squamata | Lacertidae | Gallotia | Gallotia stehlini | 2468651 |



